
Design Objectives
Our design was guided by the needs of three user groups:

Simple scientific protocols
Standardized parts
Well characterized and developed system
Standard, easy to use system
Simplified protocols
Modular tool that can be tailored to the application
Modular experiments
Open-source
Open-source
Promote good safety practices
Standardized parts
Resulting in the identification of four core design considerations for our system:

Overview
To ensure utility of our system, we developed a simplified expression and purification strategy to produce the necessary biomachinery for cell-free synthetic biology.
Our system includes all of the 36 essential TX-TL proteins, the 23S and 16S rRNA, and a tRNA for each amino acid, where tRNAPhe will act as a proof of concept for our novel purification method. In addition, we include the MS2 coat protein which is essential to our tRNA and rRNA purification strategies.

In total we have designed 41 parts! (link to parts pages)
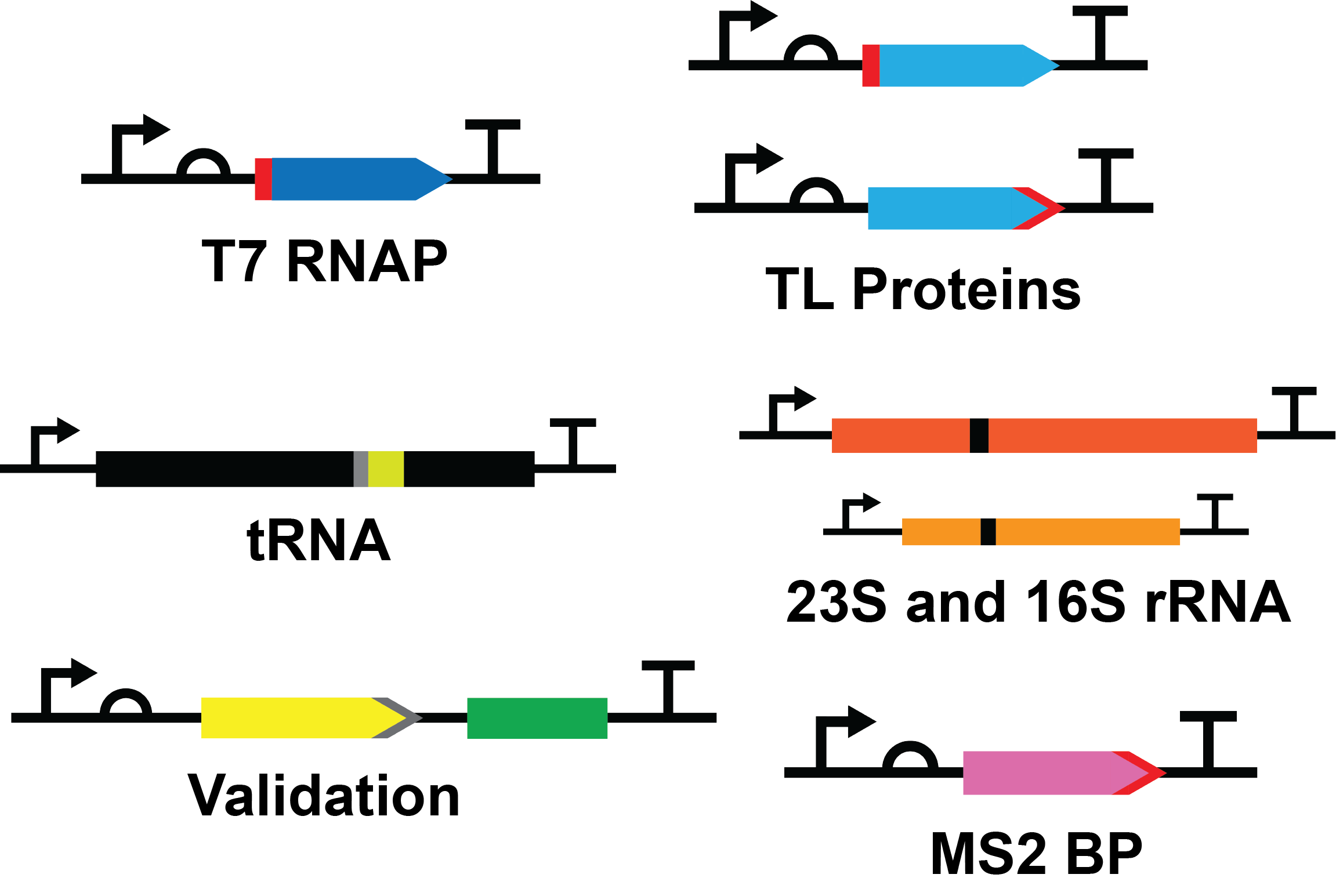
Standard Construct Design
Protein Constructs
Each protein construct consisted of a T7 promoter (BBa_I719005), medium ribosome binding site (RBS) (BBa_B0034), protein coding sequence, serine-glycine linker and histidine tag at the N- or C-terminus, and a double terminator (BBa_B0015).
The promoter, RBS and terminator are standard BioBrick parts well-characterized in the registry, providing a good starting point for the initial characterization of our system. Each construct was also codon optimized to allow for efficient expression in E. coli.

RNA Constructs
Each RNA construct consisted of a T7 promoter (BBa_I719005), RNA product, and a double terminator (BBa_B0015).

Purification Strategy
Protein Purification
Transcription and translation factors are expressed with a 6 histidine residue tag to allow for simple purification by nickel sepharose affinity chromatography. The polyhistidine tag is encoded at the N- or C-terminus of each gene based on what was previously described in the literature [1].
RNA Purification
To purify RNA, we take advantage of a previously published purification strategy adapted from the MS2 bacteriophage [2]. It takes advantage of a specific RNA: protein interaction between an RNA hairpin in the MS2 genome and the MS2 coat protein (MS2BP) that forms the viral capsid. The MS2BP has a polyhistidine tag expressed at its C-terminus. This alteration gives the MS2 coat protein dual binding ability. It will have high affinity to nickel as well as to the MS2 hairpin. Thus, tRNA and rRNA expressing an MS2 hairpin can be purified in the same column as the polyhistidine-tagged proteins.

We developed a novel purification system based on a general construct, whereby an RNA of interest will be transcribed with a scar and spacer region, acting as an oligo binding site. A DNA oligo can bind to this site, resulting in a DNA-RNA hybrid, as a target for cleavage by RNase H. This will be followed by two MS2 hairpins, enabling tight binding to the MS2BP for purification.
Our system is designed such that all components can be purified at the same time by nickel sepharose affinity chromatography!

tRNA
In nature the 20 standard amino acids are encoded in the DNA by 1 - 6 unique codon sequences, each of which has a corresponding tRNA molecule. In our system, we aim to have one corresponding tRNA for each amino acid to reduce complexity and ensure each amino acid can be incorporated.
To test the RNA purification method we have created a construct encoding for tRNAPhe. An ideal tRNA to use as it has been extensively studied compared to other tRNA molecules.

Once cell lysate containing overexpressed tRNA is added to a column containing MS2BP-Nickel Complex, the MS2 hairpins will bind to the MS2BP with high affinity. A DNA oligo complementary to the spacer and scar will be added and will anneal to this region, resulting in a short DNA-RNA hybrid region. Following oligo binding, RNase H will be added. RNase H will selectively cleave the RNA in this DNA-RNA hybrid. Subsequently, tRNAPhe with the correct 3’ end will be released into solution allowing for the isolation of tRNA from the MS2 hairpins and MS2BP by separating the supernatant from the Nickel column.

Ribosomes
The central component of translation is the ribosome. The ribosome is a large complex comprised of three ribosomal RNAs (rRNAs) and over 50 proteins in E. coli. As such, it posed a challenge in designing a simple purification strategy.
With the TX-TL components we could tag each protein with a polyhistidine tag to purify each individually. With the ribosome, tagging each ribosomal protein and the rRNA does not ensure that functional ribosomes will form after the purification. To reduce the purification of nonfunctional ribosome components we decided to utilize a previously published ribosome purification strategy that incorporated the MS2 hairpin into the rRNA genes [2].
One MS2 hairpin is inserted into both the 23S and 16S rRNA gene at regions that are surface exposed and nonfunctional in the 3D structure. This allows for a histidine-tagged MS2BP to bind the MS2 hairpin of a fully assembled ribosome to all for a one-step purification using nickel sepharose affinity chromatography. Te ensure we purify both subunits of the ribosome the MS2 hairpin will be added into the gene of both subunits rRNA gene.

Validation Construct
To validate the functionality of our TX-TL system and to provide a measure for Next vivo quality control, an additional construct was designed.

The construct consisted of a T7 promoter (BBa_I719005), a medium RBS (BBa_B0034), the coding sequence for enhanced yellow fluorescent protein (EYFP; BBa_E0030) followed by a 3X flag tag (BBa_T2004), coding sequence for the spinach aptamer [3] and a double terminator (BBa_B0015). The entire EYFP coding sequence was codon optimized for efficient expression in E. coli.
To validate successful transcription, green fluorescence is observed following addition of the fluorophore 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) which binds to the spinach aptamer mRNA.
To validate successful translation, yellow fluorescence is observed following expression of EYFP. Additionally, the 3X flag tag at the C-terminus of EYFP provides another method of protein quantification via western blot and as an internal size marker control for the expression of any other flag tagged protein.
Next vivo 2.0
Do you love the enhanced functionality and easy production of the Next vivo system? Are there features you wish Next vivo had? Well look no further than the exciting future improvements coming to Next vivo!
Advanced tRNA purification module - current tRNA purification design requires additional steps following the Ni-sepharose purification. Using a MS2BP-RNase H fusion protein that positions the RNase H near the desired cut-site allows for the tRNA to be eluted by the addition of the DNA oligo making it a one step elution as well.
Additional protein factor modules - the base Next vivo system provides all the components for basic transcription and translation, but it might not be ideal for those tricky protein or RNA products you want to make. Several additional factor sets can be added to the Next vivo system to facilitate difficult translation processes including:
- the heat shock factor package that aid in the proper folding of large or unstable protein products; or
- the orthogonal tRNA aminoacylation package that allows specific tRNA to be labeled with non-standard amino acids which are then incorporated into your protein products providing new properties to biomolecules!
Extreme temperature stability - The TX-TL components in the Next vivo system will be replaced with those from two thermophilic organisms allowing TX-TL at higher and lower temperatures with the same level of protein or RNA output!
And more coming soon!
Disclaimer: These Next vivo design features are not currently available but are some of the wild ideas and improvements that we foresee for the Next vivo TX-TL system!
References
- [1] Wang, H.H., et al., Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multi-enzyme catalysis. ACS synthetic biology, 2012. 1(2): p. 43-52.
- [2] Youngman, E.M. and R. Green, Affinity purification of in vivo-assembled ribosomes for in vitro biochemical analysis. Methods, 2005. 36(3): p. 305-12.
- [3] Strack, R.L., M.D. Disney, and S.R. Jaffrey, A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Meth, 2013. 10(12): p. 1219-1224.

