Overview
Model
Contents
Modeling is usually used to make sense of the experimental discovery in traditional biological studies. In this synthetic biology project, we believe that carefully carried out modeling will be critical for the experimental design and data analysis at different stages of the project. We hope to demonstrate that the modeling is especially helpful to finalize the microfluidics chip design, and determine the distribution of the C.elegans (Caenorhabditis elegans) in microfliudics, as well as the transfection of the MiniMos in our experiment.
Model for Chemical Diffusion
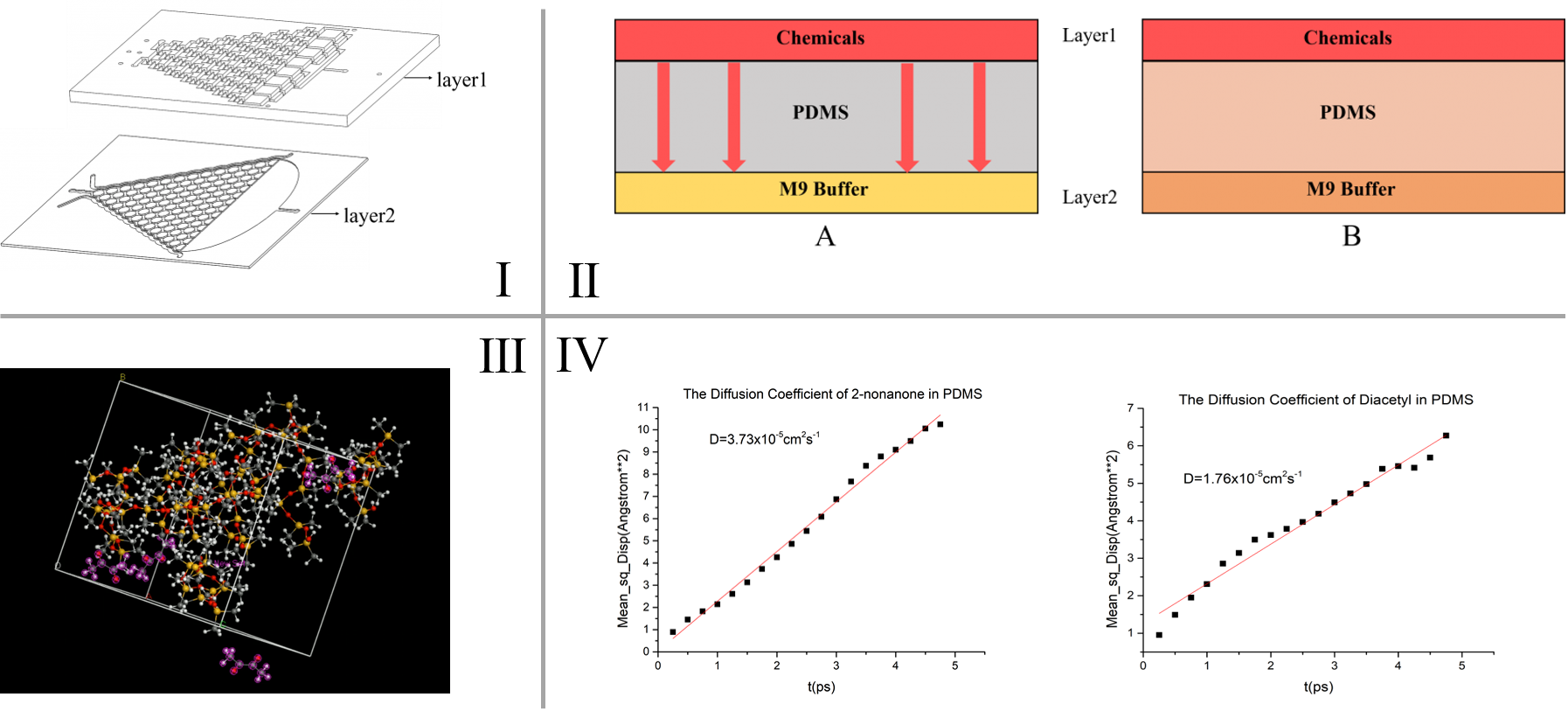
A diffusion model is built to simulate the diffusion process of chemicals in the PDMS chips, and through that we are able to determine the “delaying time” of the C. elegans’ olfactory response in our inducing experiment. The physical basis of this model is Fick’s second law (\frac{\partial C}{\partial t}=D \frac{\partial^2 C}{\partial x^2}) to know the chemical diffusion in PDMS (Polydimethylsiloxane). To calculate the time t, the diffusion coefficient D (representing the diffusion rate of chemicals in PDMS (Polydimethylsiloxane)) is needed. This parameter is calculated by simulating the force field of the diffusion of gases in silicone polymers in Material Studio®. [1] [2]
Model for Worms Locomotion
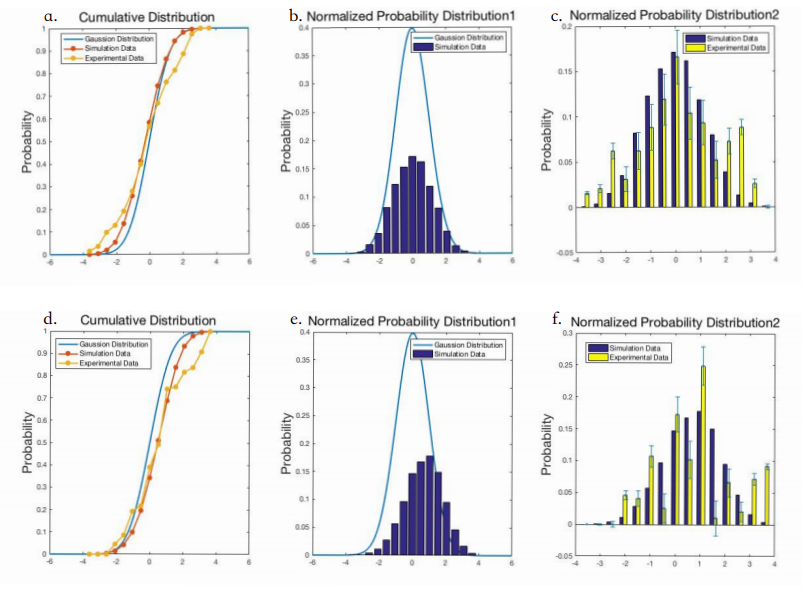
This model describes how microfluidics Gaussian distribution plate works when we use this device to test C.elegans’ preference in the plate.
Model for Neuron Network
The purpose of this model is to select promoters used in downstream neurons of C.elegans’ which have greatest possibility to be lighted on when AWA and AWB (the main neurons in our project) are actived.
Model for Genetic Probability
This model describes the process in which we get 2 worm strains with stable genetic traits and the hybrid offspring. It was proposed at the beginning of our project to optimize our experimental design concerning how many worms we need to inject and how long it will take to make sure we get the expected result before iGEM Jamboree.
These 3 steps are considered in the model:
Inject plasmid (odr-10::CoCHR::GEM-GECO::mCherry and str-1::Chrimson::GEM-GECO::GFP) through miniMOS system.
Select the rescued worms using mCherry and GFP.
Mate the 2 strains to get the hybrid offspring of red fluorescence on AWA neuron and green fluorescence on AWB neuron.
Comparing the calculation result and the experiment result, we demonstrate that the 2 strains that we finally get have the insertion gene on different chromosomes.
References
- ↑ Albrecht, D. R. and C. I. Bargmann (2011). “High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments.” Nature Methods 8(7): 599-605.
- ↑ Troemel, E. R., Kimmel, B. E., & Bargmann, C. I. (1997). Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell, 91(2), 161-9.