| Line 40: | Line 40: | ||
After getting the inserted worms, our next step is to get the hybrid by mating 2 strains. In this step, the probability of obtaining stable inheritance worms depending on the location of inserted gene on the chromosome. | After getting the inserted worms, our next step is to get the hybrid by mating 2 strains. In this step, the probability of obtaining stable inheritance worms depending on the location of inserted gene on the chromosome. | ||
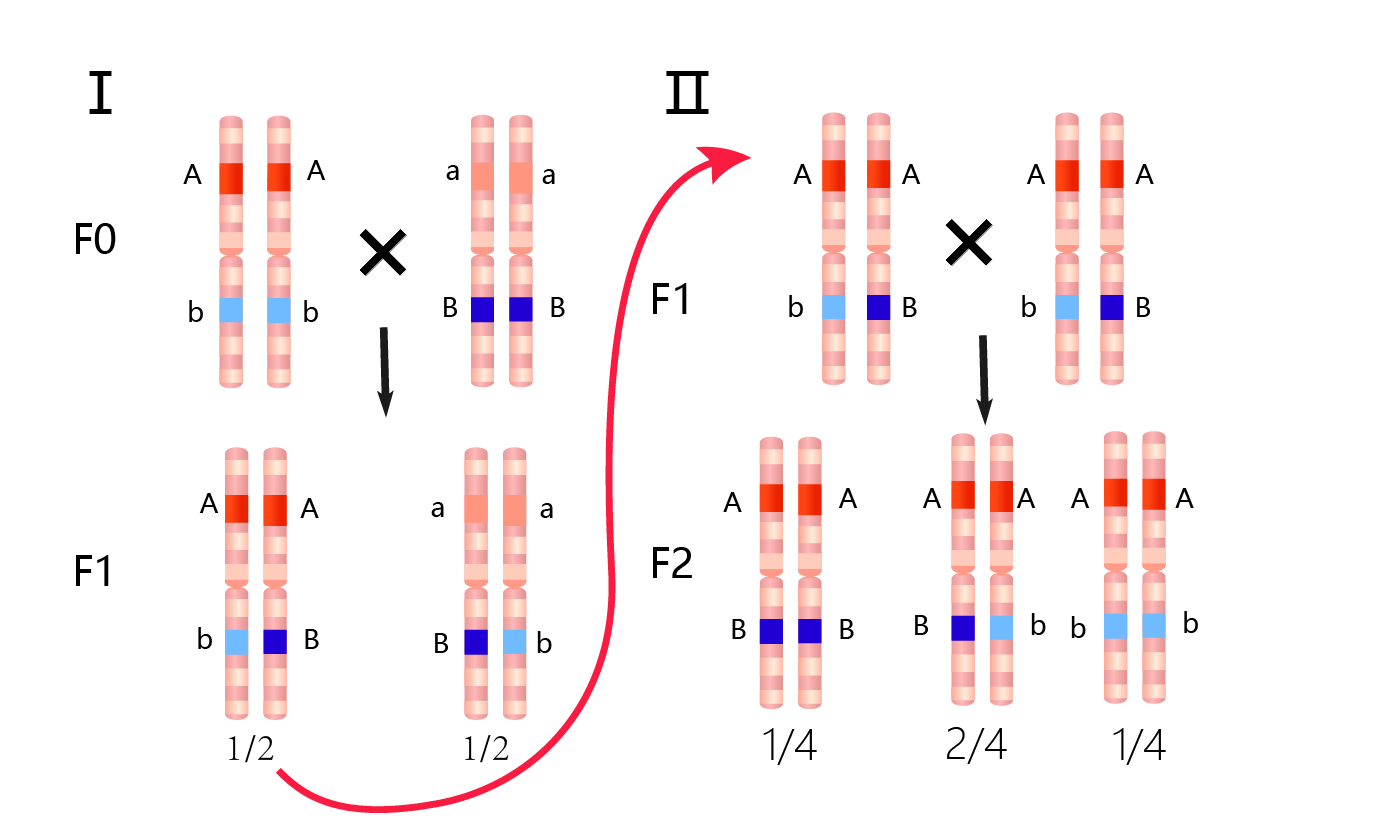
| − | {{SUSTech_Image| filename=T--SUSTech_Shenzhen--model--genetic1.png|width=450px|caption=<B>Fig 2.4 Stable inheritance probability</B> }} | + | {{SUSTech_Image| filename=T--SUSTech_Shenzhen--model--genetic1.png|width=450px|caption=<B>Fig 2.4 Stable inheritance probability</B> |
| + | F0: we use ord-10::CoCHR::GEM-GECO::mCherry as male parent and Str-1::Chrimson::GEM-GECO::GFP as female parent. | ||
| + | F1: heterozygote worms with GFP and mcherry. | ||
| + | F2: Four phenotypes of F2, AABB is what we want.}} | ||
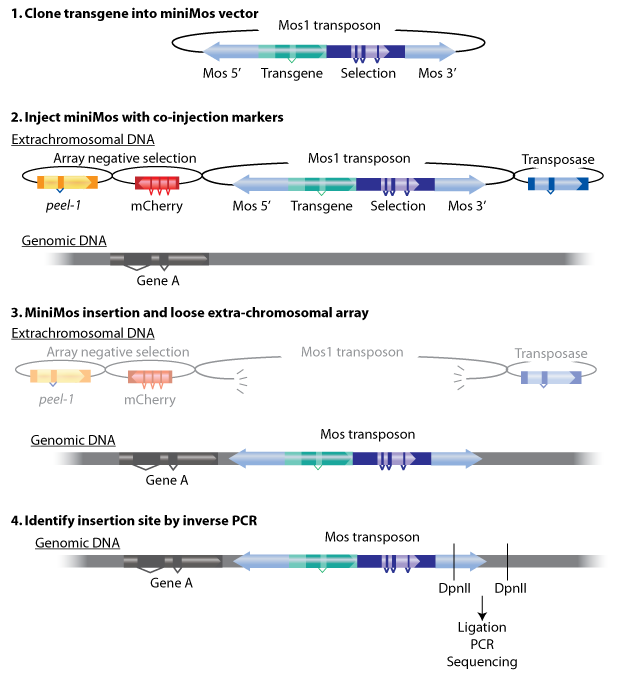
We use pCFJ909 vector as the carrier of insertion gene. It contains mos-1 transposon that can insert the gene on the chromosome randomly. Worms have 6 pair of chromosomes, so there is the probability of 5/6 that 2 strains with the inserted gene on different chromosomes and the probability of 1/6 that 2 traits are at the same chromosomes. | We use pCFJ909 vector as the carrier of insertion gene. It contains mos-1 transposon that can insert the gene on the chromosome randomly. Worms have 6 pair of chromosomes, so there is the probability of 5/6 that 2 strains with the inserted gene on different chromosomes and the probability of 1/6 that 2 traits are at the same chromosomes. | ||
| Line 52: | Line 55: | ||
If two insertions located on same chromosome, we still can get worms expressing two light-sensitive proteins by crossing over. However, the time will be much longer than the first circumstance considering the low frequency of chromosome crossing-over. | If two insertions located on same chromosome, we still can get worms expressing two light-sensitive proteins by crossing over. However, the time will be much longer than the first circumstance considering the low frequency of chromosome crossing-over. | ||
| − | {{SUSTech_Image| filename=T--SUSTech_Shenzhen--model--genetic2.png|width=450px|caption=<B>Fig 2.5 Chromosome crossing probability</B> }} In nature, the probability of crossing over never exceed 50%. And according to our experiment, if these two genes are on one chromosome, their offspring will not occurred segregation of character. So, in our situation, odr-10::CoCHR::GEM-GECO::mCherry and str-1::Chrimson::GEM-GECO::GFP are localized on two different chromosomes. | + | {{SUSTech_Image| filename=T--SUSTech_Shenzhen--model--genetic2.png|width=450px|caption=<B>Fig 2.5 Chromosome crossing probability</B> |
| + | Part I: F0: Homozygote worms obtained via microinjection. Gene A and B are localized on same chromosome. F1: heterozygote offsprings | ||
| + | Part II: Crossing over happened. F2: Three genotypes obtained in F2 | ||
| + | }} In nature, the probability of crossing over never exceed 50%. And according to our experiment, if these two genes are on one chromosome, their offspring will not occurred segregation of character. So, in our situation, odr-10::CoCHR::GEM-GECO::mCherry and str-1::Chrimson::GEM-GECO::GFP are localized on two different chromosomes. | ||
Moreover, we used our model to guide our results. We have proved that odr-10::CoCHR::GEM-GECO::mCherry is inserted on chromosome 1 by mapping( a method used to identify the locus of a inserted gene by PCR, first digest DNA into fragments, then ligate to form circular DNA and PCR to find the locus), but we failed to map the str-1::Chrimson::GEM-GECO::GFP. However, based on mating results, we can confirmed that they were inserted on different chromosomes. | Moreover, we used our model to guide our results. We have proved that odr-10::CoCHR::GEM-GECO::mCherry is inserted on chromosome 1 by mapping( a method used to identify the locus of a inserted gene by PCR, first digest DNA into fragments, then ligate to form circular DNA and PCR to find the locus), but we failed to map the str-1::Chrimson::GEM-GECO::GFP. However, based on mating results, we can confirmed that they were inserted on different chromosomes. | ||
Revision as of 02:15, 1 November 2017
Genetic Probability
Model
Genetic probability model
This model describes the process in which we get 2 worm strains with stable genetic traits and the hybrid offspring (with both the preference of blue light and the repulsion to the red light). It was proposed at the beginning of our project, so we were able to decide the frequency of microinjection and estimate how long it would take to get the result. It is a time-consuming step, and to some extent, it determines the whole experiment process.
Select the rescued worms using mCherry and GFP.
Mate the 2 strains to get the hybrid offspring of red fluorescence on AWA neuron and green fluorescence on AWB neuron.
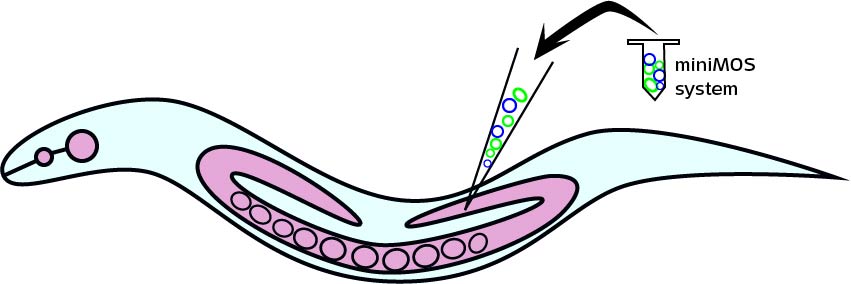
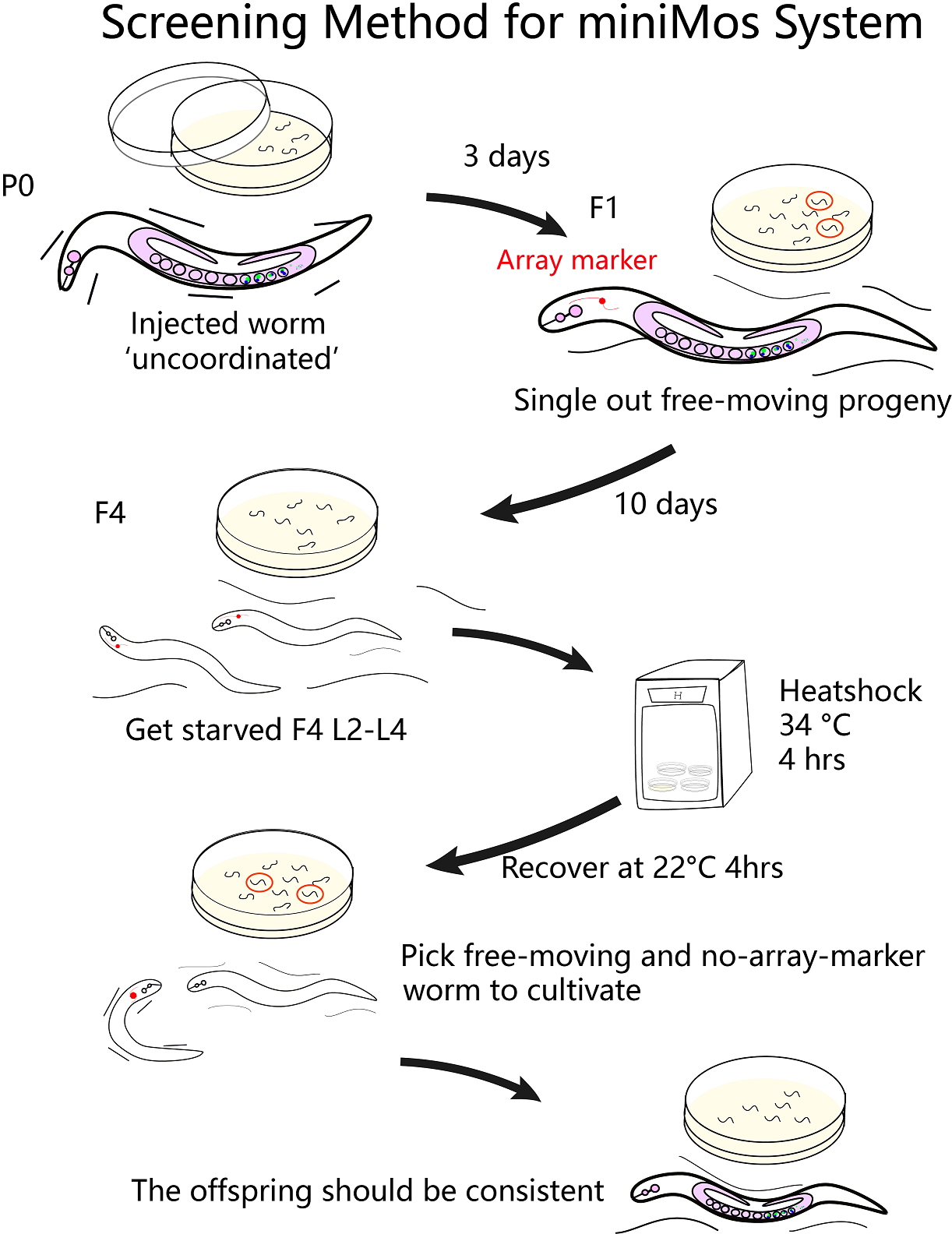
Plasmid injection and selection
To estimate the total number of worms X that we need to inject, many factors should be considered. It is said in the protocol of miniMos system, “we approximately get insertions from 60% of injected animals and from approximately 10% of all rescued F1 animals. But these numbers will depend greatly on how much practice you have in injecting. ”[1]
After long period of practice, our successful rate of injection is around 12%~25%. In calculation, we assume that the rate is 20%. After injecting the gene to its gonad, each worm produces approximately 10 embryos. About 10% of F1 containing the insertion gene can be rescued(The unc-119 gene on the vector plays a key role in ciliary membrane localization of proteins and is required for the establishment or function of the nervous system). Therefore, the number of injected worms we will get from miniMos injection is:X * 20% * 60%* 10 * 10%
Therefore, to finally get a stain, at the beginning we need to inject 9 worms at least.
After injection and selection, the total time of getting one inserted worms is about 2 weeks.
In fact, we inject totally 40 worms at 2 times to get the odr-10::CoCHR::GEM-GECO::mCherry insertion, and 40 worms to get the str-1::Chrimson::GEM-GECO::GFP. The total time consumption is one and a half months since we injected worms separately.
Mating for stable inheritance
After getting the inserted worms, our next step is to get the hybrid by mating 2 strains. In this step, the probability of obtaining stable inheritance worms depending on the location of inserted gene on the chromosome.
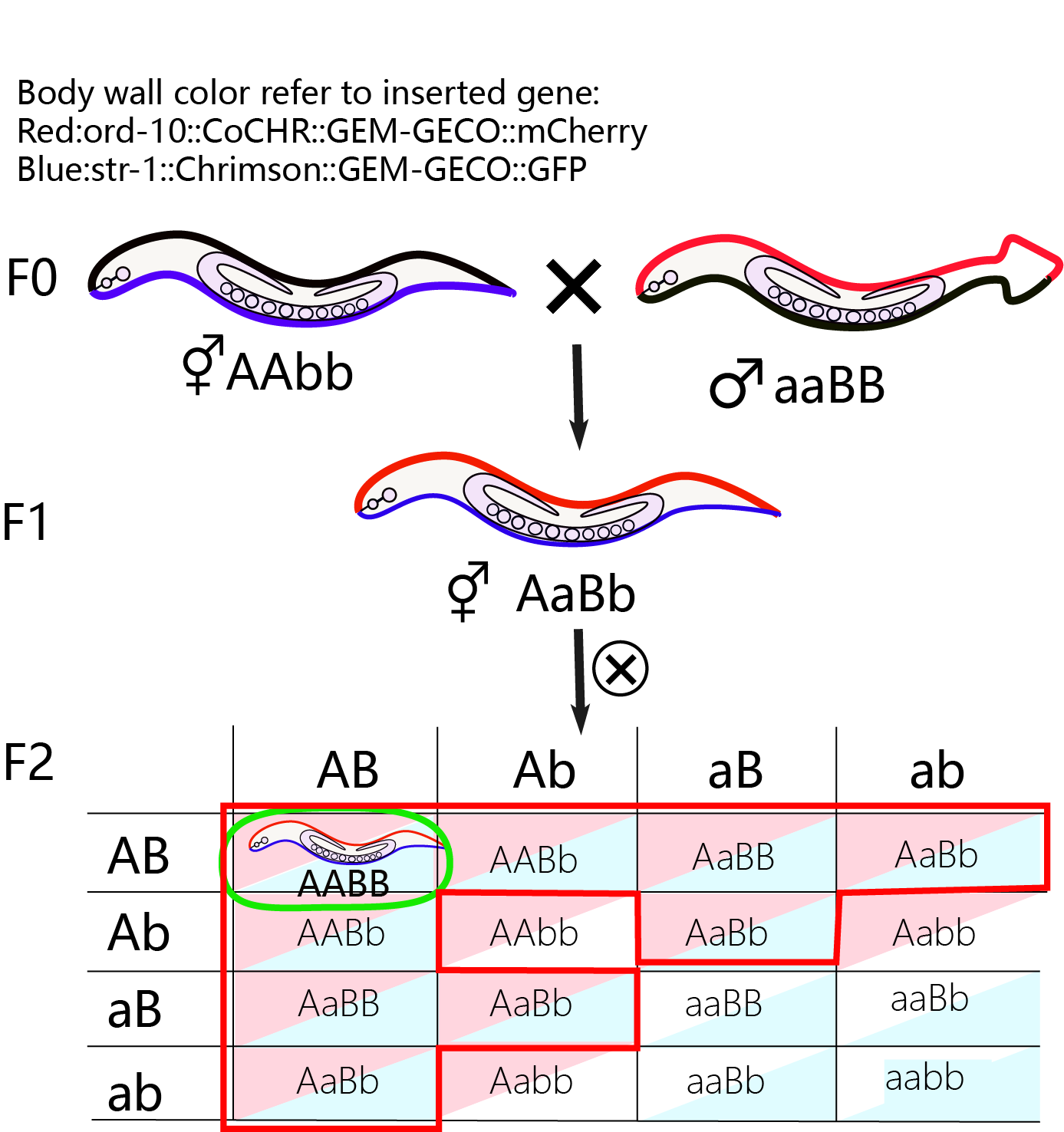
1) Different chromosomes insertions: The probability to get stable inheritance is based on the Mendel’s laws. Basically, F1 will has both of 2 traits and there will be trait segregation in F2. 9/16 of F2 worms will contain RFP and GFP in two different neurons. Then we continue to exam whether there will be trait segregation among F3. The number of worms with the stable inheritance of 2 traits consist 1/16 of the total number of F2 worms. In this step, the time cost is about 2 weeks.
2) Single chromosome insertion:
If two insertions located on same chromosome, we still can get worms expressing two light-sensitive proteins by crossing over. However, the time will be much longer than the first circumstance considering the low frequency of chromosome crossing-over.
Moreover, we used our model to guide our results. We have proved that odr-10::CoCHR::GEM-GECO::mCherry is inserted on chromosome 1 by mapping( a method used to identify the locus of a inserted gene by PCR, first digest DNA into fragments, then ligate to form circular DNA and PCR to find the locus), but we failed to map the str-1::Chrimson::GEM-GECO::GFP. However, based on mating results, we can confirmed that they were inserted on different chromosomes.
After we did mating experiments and among 40 F2 generation, two of F2 worms showed stable GFP and RFP in their offspring, which obeys the Mendel’s laws.
It cost one month for us to finally get the hybrid worm strain.
References
- ↑ http://www.wormbuilder.org/minimos/commentsfaq/