Template loop detected: Template:HFLS H2Z Hangzhou<!DOCTYPE html>
Genetic Circuit
Overview
We constructed several genetic circuit either to act as sensors or act as expression device. Through our visit to Qiumei's Lab in Hangzhou, we gained priceless data concerning the salt content of their products. You have already seen nitrite natural decomposition rate, we will use that information as working environment for our several sensors. We also will include the nitrate content information in their products to be used as working environment for our several sensors. We constructed and standardized several expression device as well, some we tested, some we didn't. By submitting parts to registry, we hope latter years' iGEM teams can further our research and pick up some of our characterization to be used in their own design.
Sensors
We constructed in total two sensors, all start with the nitrite/nitrate responsive promoter Pyear on the right. We also tested one basic sensor Part:BBa_K381001 and improve characterization of the part. In fact, the new sensor Part:BBa_K2346006 is a functional improvement to K38001.Protocols for measuring the sensors
We use a set of protocols to measure the strength of these sensors so we could cross-reference every sensors with each other.Procedures for making sample to be measured its fluorescence
1 |
Select single colony from plate, place in LB culture with CmR resistance (50μg/mL), 37°C culture 12 hours. |
2 |
Placed in 200μL M9 culture, 250rpm, 37°C. Making the final bacteria diluted 50 times. |
3 |
Dilute the bacteria in step 2 20 times with M9 culture, remove 100μL to be added in the 96 well plate. Using samples without addition of nitrogen-containing salt as control, and M9 as blank. |
4 |
End the program after 4 hours. |
Protocol for measuring Fluorescence Intensity
| Plate Type: 96 WELL PLATE (Use plate lid) | |
| Eject plate on completion | |
| Set Temperature: Setpoint 37°C, Gradient 0 °C Preheat before moving to next step |
|
| Read: Absorbance Endpoint Full Plate Wavelengths: 600 Read Speed: Normal, Delay: 100 msec, Measurements/Data Point: 8 |
|
| Read: Fluorescence Endpoint Full Plate Filter Set 1 Excitation: 485, Emission: 528 Optics: Top, Gain: 100 Light Source: Xenon Flash, Lamp Energy: High Read Speed: Normal, Delay: 100 msec, Measurements/Data Point: 10 Read Height: 7 mm |
|
| Start Kinetic: Double Orbital: Continuous Frequency: 282 cpm (3 mm) |
|
| Read: Absorbance Endpoint Full Plate Wavelengths: 600 Read Speed: Normal, Delay: 100 msec, Measurements/Data Point: 8 |
|
| Read: Fluorescence Endpoint Full Plate Filter Set 1 Excitation: 485, Emission: 528 Optics: Top, Gain: 100 Light Source: Xenon Flash, Lamp Energy: High Read Speed: Normal, Delay: 100 msec, Measurements/Data Point: 10 Read Height: 7 mm |
Background
This is a reporting device/sensor made by team iGEM10_BCCS-Bristol, the construct is a normal GFP reporter gene attached to the PyeaR promoter, responsive to nitrate, nitrite and nitric oxide.Results
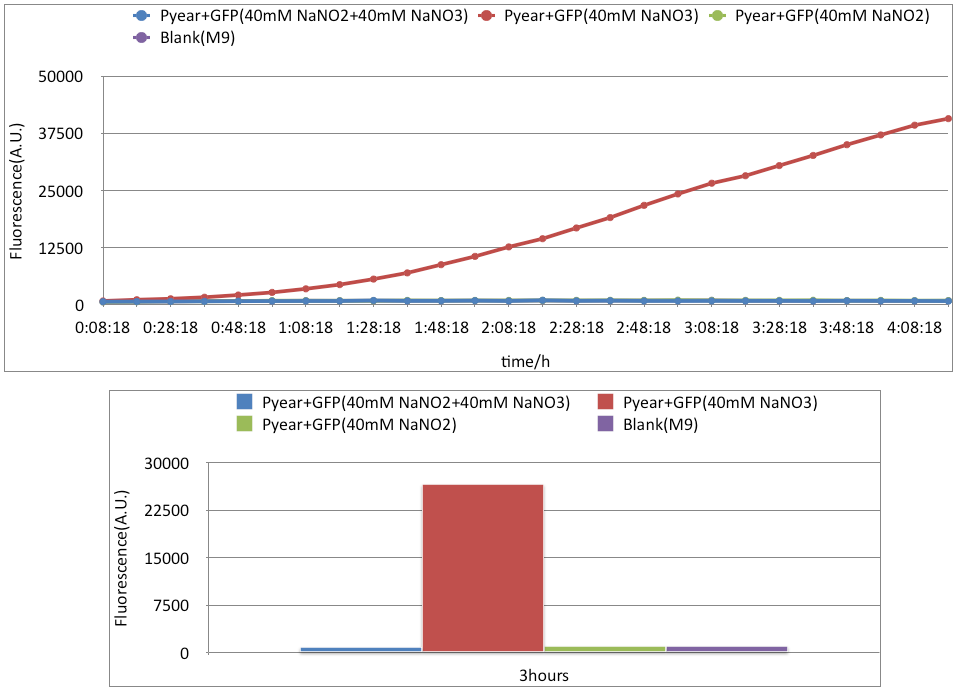
Conclusion
In our experience with this part, we found out that Pyear will yield strong transcription rate under the presence of high concentration of Nitrate. However, we found out, under high concentration of nitrite(40mM), the promoter will be repressed, unlike nitrite in low concentration. Furthermore, if nitrate(40mM) is present along with nitrite(40mM), the promoter will be inhibited.Background
Nitrate/nitrite responsive promoter with amplifierThis is a composite part and a functional improved device for Part:BBa_K216005. The Pyear promoter will be switched on under the presence of nitrate or nitrite, translating P2 ogr. The P2 ogr will cause inducible PF promoter to produce even stronger downstream PoPS. We attached a RBS for convince of registry users.
We ligated this part with a GFP+terminator.
Design
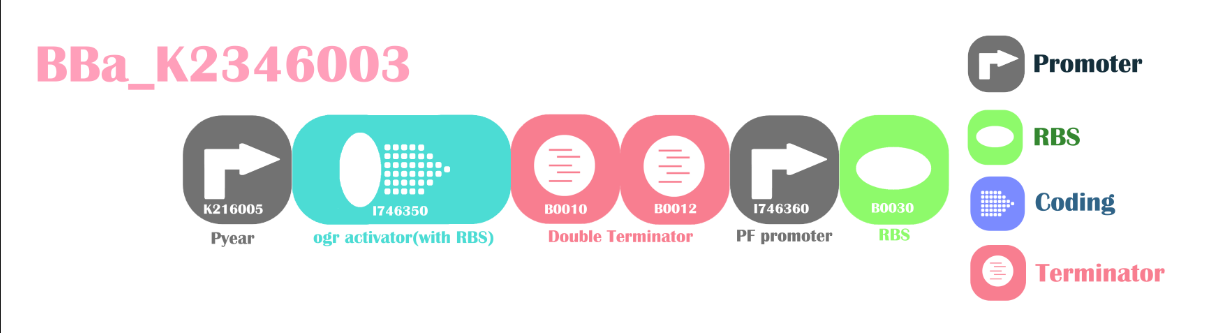
Results
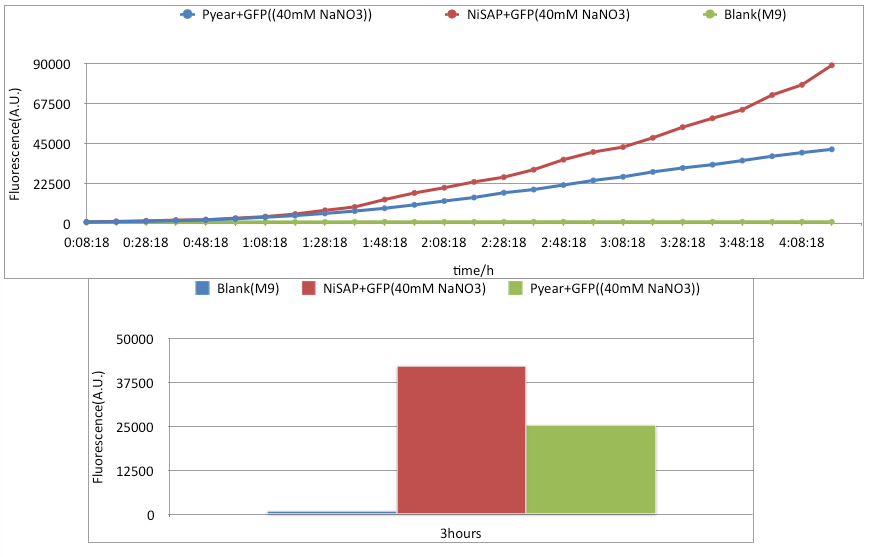
Conclusion
As we can see from the graph, there is significant difference between NiSAP's response to nitrate(40mM) and Pyear+GFP's response to nitrate(40mM). After 3 hours in the plate reader, NiSAP's A.U. is nearly twice the magnitude of Pyear+GFP.Background
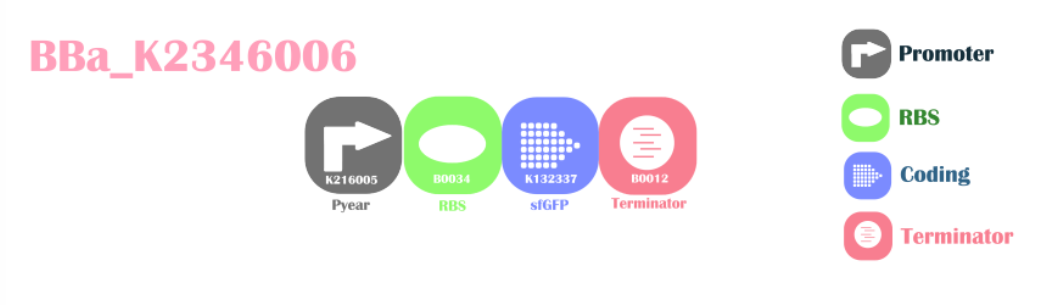
Enhanced Sensor for Nitrite/NitrateThis is a functional improved device for Part:BBa_K381001. We replaced the original reporter gene GFP with brighter reporter gene sfGFP, in the hope to improve the sensitivity for the device to detect nitrite and nitrate.
Design
Results
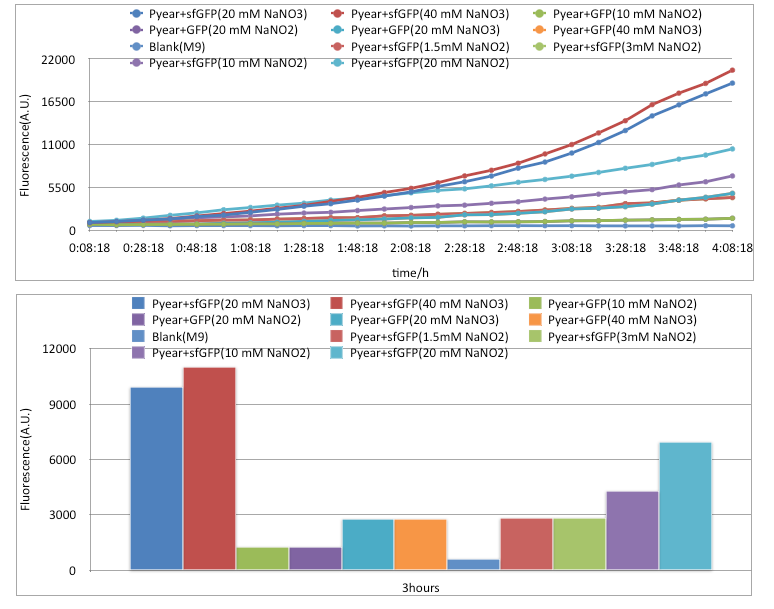
Conclusion
We tested several environment for Pyear+sfGFP and compared it with Pyear+GFP. Our device compared with previous reporter Part:BBa_K381001 is significantly better in every aspect. Furthermore, the sensitive towards nitrite is greatly improved: just with a concentration of 1.5mM nitrite, sfGFP can give equal A.U. value compared with GFP device in 40mM nitrate. Furthermore, using sfGFP, we can draw distinction between 40mM and 20mM nitrate, previously not feasible with Pyear+GFP.Expression Device
We constructed several genetic circuit expressing proteins. We submitted composite parts with part number BBa_K2346003 to BBa_K2346007 to the registry. See our performance(https://2017.igem.org/Team:HFLS_H2Z_Hangzhou/Demonstrate) for more information.BBa_K2346004
Background
Nitrate/nitrite responsive promoter with autoinducer synthetase for AHL(3OC6HSL)This is a composite part. The Pyear promoter will be switched on under the presence of nitrate or nitrite, translating LuxL protein. LuxL is an enzyme for catalyzing AHL(acyl-homoserine lactones) from normal cell metabolites.
Design
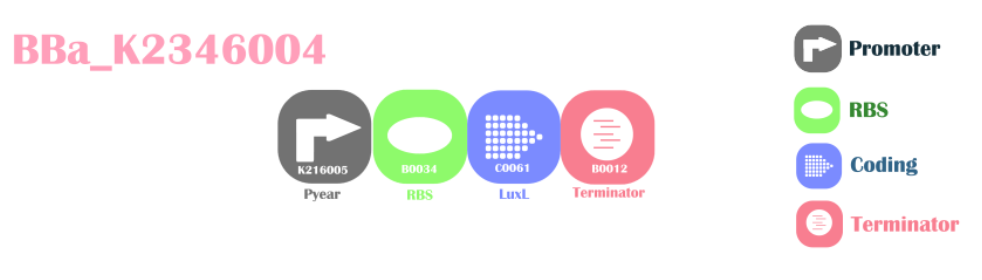
BBa_K2346005
Background
Positive Feedback Loop for NzFEThis is a device to reduce nitrite ion to its elements: oxygen and nitrogen. The device is regulated by signaling molecule AHL(3OC6HSL). The right p(tetR) promoter will constitutively express Plux’s repressor: LuxR protein. Two LuxR protein molecules can bind with two AHL molecules to form a complex which in turns switches on the PluxR promoter. The down stream LuxL is an enzyme for catalyzing AHL(acyl-homoserine lactones) from normal cell metabolites. The LuxL translated will increase the AHL content in the chassis. This forms a positive feedback loop with increasing AHL resulting in increasing PoPS from PluxR. We utilize this increasing PoPS, and place CDS for NzFE(Nitrite Degradation Fusion Enzyme) in the down stream of the circuit.
Design
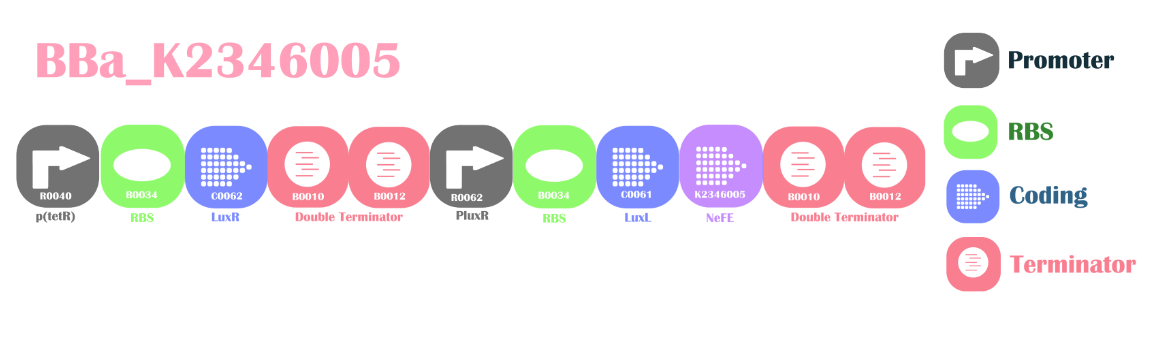
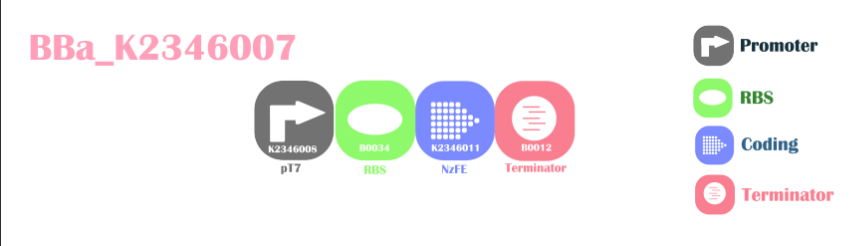
BBa_K2346007
Background
NzFE T7 expression vectorThis is a composite part for expression of NzFE in DE-type bacteria. The pT7 promoter can only be used in chassis with T7 RNA synthase. Though pT7 commonly thought to be constitutive, we encountered the problem of unable to express the protein without IPTG (lactose replacement). We suspect the company providing BL21 DE strand competent cell changed the genome of their bacteria, made T7 RNA synthase regulated by IPTG.
Design