The Problem
Co-cultures are fast becoming more widely researched, including with respect to industrial applications. In order to fully understand and, thence, optimise the growth of organisms in co-culture, it would be helpful to study that growth directly. In other words: to be able to monitor the growth rates of the individual constituents of a co-culture, as well as the total growth rate, without disturbing the system. There exist several methods of doing this, currently, though all have their shortcomings (as explained below). We designed our hardware and software with the intentions of distinguishing between organisms in situ and obtaining cell counts for a whole sample and each of its constituents. We feel that this offers at least some improvement over the existing analysis methods.
Existing Analysis Methods
- Plating
Growing organisms on plates is perhaps the most rudimentary analysis technique for studying growth rates and the interactions between organisms. The process is time consuming since the formation of colonies for most organisms takes days. Further, if colony counting is performed by hand (rather than using some software or other tool) yet more time is taken to obtain results. Moreover, the number of colony forming units found by colony counting is not necessarily representative of the number of organisms with which the plate was inoculated. There is no way of reliable ascertaining information about the intermediary growth stages of the organism(s) in question using this method.
- Staining & Optical Microscopy
While staining and observing cells under an optical microscope is not so time consuming as the above plating method, it has its own limitations. In order to prepare samples and record data, someone must be present at all times. This is not true of the design of our Quicker Way to Analyse Co-Cultures (QWACC) system. (That is, the combined upright DIHM and milli-fluidic chamber.) Although, as our Results suggest, we had some trouble getting everything to work simultaneously, we firmly believe that the design is capable of carrying out automated co-culture analysis with relatively little change. Further, we need not remove samples from our cultures or stain them, meaning a less time-consuming process with fewer opportunities for contamination and no need for a person's constant presence.
- Haemocytometer & Automated Cell Counter
We used this as the gold-standard for accuracy in our tests. We compared our results with the results from a ThermoFisher Countess II FL Automated Cell Counter. However, the machine we used was not able to differentiate between organisms in a co-culture without fluorescence and, at £4070 [1], is far more expensive than our DIHM, which cost around £150 to build. Of course, it is often possible to insert a fluorescence gene into a given organism, which would allow for the use of the Automated Cell Counter. Though, this is not always possible and, even when it is, means altering the organisms being studied. Holographic microscopy does not require fluorescence to distinguish organisms and so can be used with wild-type organisms.
- Other Techniques Using Fluorescence
There are many techniques that could use fluorescence to calculate the relative densities of organisms in co-cultures. These include, but are not limited to, flow cytometry and fluorescent microscopy. As stated above, our technique does not require that the organisms in a culture fluoresce. This drastically improves the versatility of the technique with respect to the variety of organisms on which it can be performed. Further, we need not dispose of samples after they have passed through the analysis chamber. This means, as was stated with reference to staining and optical microscopy, that there are fewer opportunities for contamination.
- Metabolic Rate Analysis
This method is cheap, not too time consuming and is non-disruptive - samples need not be taken out of the culture. The idea is that the amounts of products of metabolic processes can be analysed and the growth of each organism that gives a different product can be inferred. For a given pair of organisms, however, it is not likely that their metabolic products are so different that the co-culture could be analysed in this manner. Also, the metabolic rates of organisms can be affected by a great number of variables, meaning that the accuracy of the technique is somewhat limited.
- Spectrophotometry
This method is more useful for finding the total cell count of a sample and, even then, it is not renowned for its accuracy, due to the fluctuations in absorbance intrinsic in a biological sample. The absorbance at different wavelengths may be useful for calculating the cell densities of the constituents of a co-culture sample, in the same manner as a pulse-oximeter is used to calculate the ratio of oxygenated to deoxygenated blood in medicine, though this is purely speculation on our part.
QWACC
We have shown, on the Results page, that by using a DIHM based analysis system, organisms in a co-culture can be differentiated between as long as they are either different shapes or different sizes. Of particular interest is the data we have shown regarding a co-sample of Chlamydomonas reinhardtii and Galdieria Sulphuraria. This serves as proof of concept. We can differentiate between organisms based on a difference in size on the order of a few microns. Obviously, there remains the possibility that the deviation in size of a single species may exceed this limit, though the technique remains promising. This is an improvement over most existing analysis methods if only because no genetic modification (fluorescence) or staining is required. Additionally, other advantageous properties of the QWACC method come from using the milli-fluidic chamber. Again, this requires improvement in practice, but, by using the milli-fluidic chamber, we propose that contamination is more easily avoidable and that continual analysis can take place with much less human input and/or supervision.
In short, the QWACC method of analysis is more versatile, more affordable and requires less work than the traditional methods of co-culture observation and it seems, at least from our results, to be similarly accurate.
Design for a Future Application
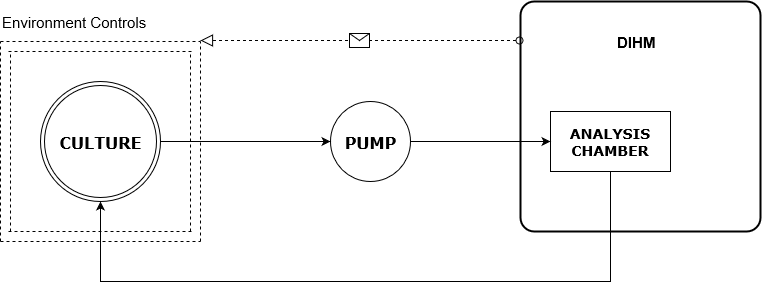
Figure 1: Feedback loop to optimise growth.
The flow chart in figure 1 shows a liquid culture in some container that regulates the environment that the organisms experience (e.g. temperature, pH, stirring, et cetera). The culture is pumped into the milli-fluidic chamber and is then analysed by the DIHM. The Raspberry Pi in the DIHM can then be used to calculate what should be done to the environment in order to optimise the growth of each of the organisms in the co-culture, using some mathematical model. This could also be done by an external computer if more processing power is required. This information is fed back into the environmental control system and the culture continues to cycle. Hence, we have a continuous analysis and optimisation system.
While we were not able to build a setup such as this in the short time we worked on this project, we believe it would be useful in an industrial context. A co-culture whose growth has been optimised for the production of some end-product would be useful, for example, in bioethanol production. There are several steps to follow before this possibility is realised: a thinner analysis chamber, a reliable mathematical model and a continuous version of the analysis software. The milli-fluidic chamber and software designs are easily manipulated. The mathematical model would have to be calculated for each given co-culture, so this would have to be done on a case by case basis.
References
- [1]: Countess II FL Automated Cell Counter, ThermoFisher Scientific, as available on 31st October 2017 at www.thermofisher.com/order/catalog/product/AMQAF1000

