| (30 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<!-- {{HK_SKHLPSS}} --> | <!-- {{HK_SKHLPSS}} --> | ||
| − | + | = Design = | |
| − | + | Since our previous studies only focused on 2-dimensional structure of nano-device, we would like to explore the feasibility of a 3-dimensional structure of nano-device. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | [[File: Design_1.jpg |thumbnail| 800px | center | Fig.3. The nano-cube is designed according to Watson & Crick’s principle. Under guanine-riched sequence, it is able to bind with specific target strand to form a G-quadruplex. The G-quadruplex will enhance the peroxidase activity of hemin and lead to the peroxidation of 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) giving a colorimetric signal. The degree of peroxidase activity indicates the concentration of the target.]] | |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| + | In our design, eight single-stranded DNAs are used to form a DNA Nano-cube. This cube is designed so that one side of the cube is originally open. We fit in the corresponding base pairs of this side so that the presence of the target H3N2 virus strands would induce the closing of the cube and lead to the formation of G-quadruplex. | ||
| + | |||
| + | The eight oligos we chose and the nano-cube we designed are listed below as reference. | ||
| + | |||
| + | <html> | ||
<table class="table table-hover table-bordered" style="font-size: 1em"> | <table class="table table-hover table-bordered" style="font-size: 1em"> | ||
| − | |||
<tr> | <tr> | ||
| − | <th style="text-align: center" width=" | + | <th style="text-align: center" width="25%">Cubic forming Oligo</th> |
| − | <th style="text-align: center">Sequence</ | + | <th style="text-align: center">Sequence</th> |
| − | <th style="text-align: center" width="15%">Size (nucleotide)</ | + | <th style="text-align: center" width="15%">Size (nucleotide)</th> |
</tr> | </tr> | ||
| Line 81: | Line 76: | ||
</tbody> | </tbody> | ||
</table> | </table> | ||
| + | </html> | ||
| − | + | The colours show how the oligos bind with each other. We used DNA sequence mimicking the RNA sequence to do the test first because of stability problem. | |
| − | + | ||
| − | + | ||
| + | <html> | ||
<table class="table table-hover table-bordered" style="font-size: 1em"> | <table class="table table-hover table-bordered" style="font-size: 1em"> | ||
| − | |||
<tr> | <tr> | ||
| − | <th style="text-align: center" width="20%">Oligo</ | + | <th style="text-align: center" width="20%">Oligo</th> |
| − | <th style="text-align: center">Sequence</ | + | <th style="text-align: center">Sequence</th> |
| − | <th style="text-align: center" width="15%">Size (nucleotide)</ | + | <th style="text-align: center" width="15%">Size (nucleotide)</th> |
</tr> | </tr> | ||
| Line 99: | Line 93: | ||
<td style="text-align: center">36</td> | <td style="text-align: center">36</td> | ||
</tr> | </tr> | ||
| − | + | </table> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</html> | </html> | ||
| − | + | After obtaining positive result in using DNA sequence, we would also like to check whether our design could successfully bind with RNA of H3N2 virus. Hence, the above RNA sequence of H3N2 virus was ordered for further study. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | The above DNA and RNA strand are sponsored by <I>Integrated DNA Technologies</I>. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{HK_SKHLPSS_FOOTER}} | {{HK_SKHLPSS_FOOTER}} | ||
Latest revision as of 12:49, 1 November 2017
Design
Since our previous studies only focused on 2-dimensional structure of nano-device, we would like to explore the feasibility of a 3-dimensional structure of nano-device.
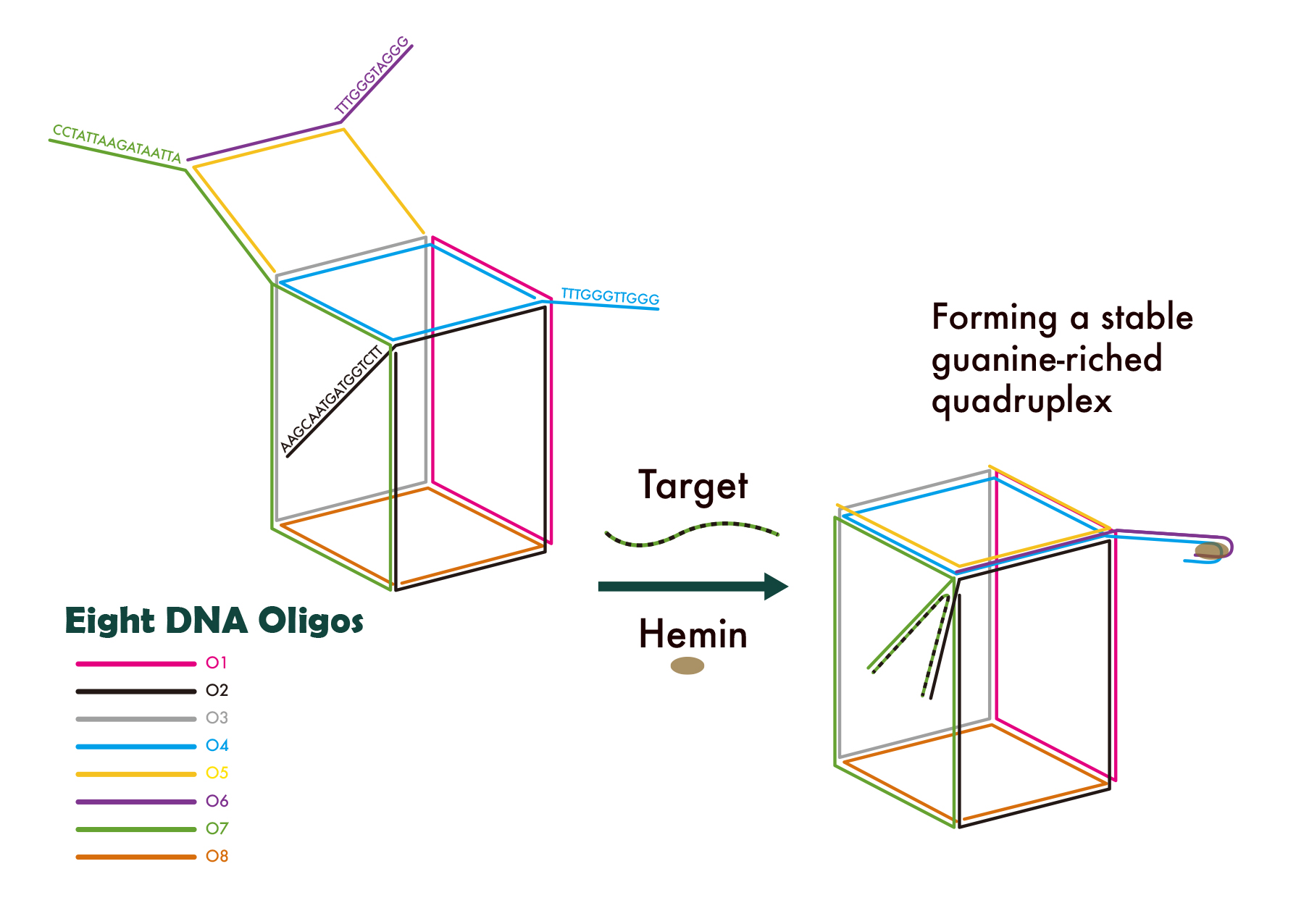
In our design, eight single-stranded DNAs are used to form a DNA Nano-cube. This cube is designed so that one side of the cube is originally open. We fit in the corresponding base pairs of this side so that the presence of the target H3N2 virus strands would induce the closing of the cube and lead to the formation of G-quadruplex.
The eight oligos we chose and the nano-cube we designed are listed below as reference.
| Cubic forming Oligo | Sequence | Size (nucleotide) |
|---|---|---|
| Oligo1 | TCG GCC GCG CGG TTC CGG TGC CCC CTT CGG CGC CCG TCT TGC GGT CCG GGG TTG CAG GAC GCC GT | 65 |
| Oligo2 | AAG CAA TGA TGG TCT TTT TCC GCG GGC CCG TTC CCC GGA CCG CTT GGG CAG CCT GGT TCC AGC CAC GGC T | 70 |
| Oligo3 | TCG CCG GGT GGG TTG AGT GGC GCC CTT GGG GGC ACC GGT TGG CAG GCG GCG T | 52 |
| Oligo4 | GGG TTG GGT TTC GGG CCC GCG GTT GCC GCG GCA GGT TCG CCG CCT GCC TTC GGC GTC CTG CT | 62 |
| Oligo5 | TCC GCG CGG CCG TTT AGG CCC GGC GTT CCC CGG CCG CCT | 39 |
| Oligo6 | TCG CCG GGC CTA TTT GGG TAG GG | 23 |
| Oligo7 | TCC TGC CGC GGC TTG CCG TGG CTG GTT CGC CGC GGC CCT TCC CAC CCG GCG TTG GCG GCC GGG GTT TAT TAA TAG AAT TAT CC | 83 |
| Oligo8 | TGG GCC GCG GCG TTC CAG GCT GCC CTT GAC GGG CGC CGT TGG GCG CCA CTC T | 52 |
| Target | GGA TAA TTC TAT TAA TCA TGA AGA CCA TCA TTG CTT | 36 |
The colours show how the oligos bind with each other. We used DNA sequence mimicking the RNA sequence to do the test first because of stability problem.
| Oligo | Sequence | Size (nucleotide) |
|---|---|---|
| Target (RNA) | GGA UAA UUC UAU UAA UCA UGA AGA CCA UCA UUG CUU | 36 |
After obtaining positive result in using DNA sequence, we would also like to check whether our design could successfully bind with RNA of H3N2 virus. Hence, the above RNA sequence of H3N2 virus was ordered for further study.
The above DNA and RNA strand are sponsored by Integrated DNA Technologies.

