Ianoverman (Talk | contribs) |
|||
| (84 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Austin_UTexas}} | {{Austin_UTexas}} | ||
<html> | <html> | ||
| + | <style> | ||
| + | body { | ||
| + | background-color: #282828 | ||
| + | } | ||
| − | + | </style> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<div class="column full_size"> | <div class="column full_size"> | ||
| − | < | + | <div style="background-color: lightcyan;padding:14px; font-family: verdana"> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | <h2 style="font-family: verdana"> Interlab Measurement Study </h2> |
| − | </ | + | <br> |
| + | <p style="font-family: verdana">The 2017 Interlab Study is the fourth such initiative by the Measurement Committee of iGEM Foundation to establish a hardy measurement procedure for the green fluorescent protein. The goal for the study this year is to test various RBS devices with the intention of finding which combinations yield more stable gene expression and how this might translate to various labs worldwide. Detailed below are the protocols and materials we used during the course of the experiment as provided by the iGEM Foundation. We are proud to contribute to this research endeavor by the iGEM Foundation.</p> | ||
| − | |||
<br> | <br> | ||
| − | <p>The | + | |
| + | <p style="font-family:verdana">The University of Texas at Austin iGEM team is one of nine teams to have participated in all of the interlab studies to date!</p> | ||
<br> | <br> | ||
<hr> | <hr> | ||
<br> | <br> | ||
| − | <h3>Protocol</h3> | + | <h3 style="font-family: verdana">Protocol: Getting Started</h3> |
</html> | </html> | ||
| − | + | <ol style="font-family: verdana"> | |
| − | + | <li>Transformed Interlab 2017 Part kit devices into <i>E. coli</i> DH5-alpha cells using electroporation.</li> | |
| − | + | <li>Plated on LB/CAM and allowed to grow overnight at 37° C.</li> | |
| − | + | <li>Two colonies from each plate used to inoculate 5 ml of LB/CAM liquid media and grown overnight at 37° C and 220 rpm for a total of sixteen culture tubes.</li> | |
| − | + | <li>Next dilutions were made in 12 ml of LB/CAM to a target OD<sub>600</sub> of 0.02 and allowed to grow at 37° C and 220 rpm in a foil covered incubator with the lights off.</li> | |
| + | </ol> | ||
| + | |||
<html> | <html> | ||
<br> | <br> | ||
| − | <h3>Plate Reader | + | <h3 style="font-family: verdana">Protocol: Plate Reader</h3> |
</html> | </html> | ||
| − | + | ||
| − | + | <ol style="font-family: verdana"> | |
| − | + | <li>Allow OD<sub>600</sub> dilutions to grow for 6 hours at 37° C and 220 rpm</li> | |
| + | <li>Take measurements every two hours, starting with hour 0, for a total of four measurement points.</li> | ||
| + | <li>Measurements were taken by:</li> | ||
| + | |||
| + | <ul style="font-family: verdana"> | ||
| + | <li>Taking 500<span>µ</span>l from each of the sixteen culture tubes and put into microcentrifuge tubes.</li> | ||
| + | <li>Next, 100<span>µ</span>l from these samples were pipetted into the wells of a Costar black with clear-bottom 96 well plate.</li> | ||
| + | <li>The samples were measured using an Infinite 200 Pro with the following settings: Excitation 485nm with a 9nm bandwidth and an Emission of 530nm with a bandwidth of 20nm. A gain of 48 was also used.</li> | ||
| + | </ul> | ||
| + | </ol> | ||
| + | |||
<html> | <html> | ||
<br> | <br> | ||
| − | <h3>Ludox | + | <h3 style="font-family: verdana">Ludox and Fluorescein Measurements</h3> |
| + | |||
| + | <p style="font-family: verdana">LUDOX-S40, provided by the iGEM foundation, was used to create a point of reference between the fluorescence data and OD<sub>600</sub>. Additionally, the provided fluorescein was used to create a standard curve of the fluorescence data gathered from the cellular measurements. </p> | ||
</html> | </html> | ||
| − | # | + | |
| − | # | + | <ol style="font-family: verdana"> |
| + | <li>LUDOX-S40 was measured by loading four, 100<span>µ</span>l samples into a plate in wells A1, A2, A3, and A4, while 100<span>µ</span>l diH<sub>2</sub>O was added to B1, B2, B3, and B4.</li> | ||
| + | <li>Fluorescein was measured by diluting 2X fluorescein with PBS to 1X and brought to a total volume of 1 mL</li> | ||
| + | <li>100<span>µ</span>l of PBS were loaded into the C1-F12 area on the same 96-well plate used for the LUDOX measurements. </li> | ||
| + | <li>Serial dilutions were performed with 100<span>µ</span>l of 1X fluorescein added to C1, mixed well, and then 100<span>µ</span>l taken from that well and put into the next well, C2. This was continued to well C11, leaving C12 with just 100<span>µ</span>l of PBS.</li> | ||
| + | <li>The above procedure was repeated for rows D-F.</li> | ||
| + | </ol> | ||
| + | |||
<html> | <html> | ||
| − | + | <h3 style="font-family: verdana">Devices Measured</h3> | |
| − | <h3> | + | |
</html> | </html> | ||
| − | + | ||
| − | + | <p style="font-family: verdana">All parts were located on the pSB1C3 plasmid backbone. The parts used as designated by the provided protocol were:</p> | |
| − | + | ||
| + | <ol style= "font-family: verdana"> | ||
| + | <li>Positive Control - [http://parts.igem.org/Part:BBa_I20270 BBa_I20270]</li> | ||
| + | <li>Negative Control - [http://parts.igem.org/Part:BBa_R0040 BBa_R0040]</li> | ||
| + | <li>Test Device 1 - [http://parts.igem.org/Part:BBa_J364000 BBa_J364000]</li> | ||
| + | <li>Test Device 2 - [http://parts.igem.org/Part:BBa_J364001 BBa_J364001]</li> | ||
| + | <li>Test Device 3 - [http://parts.igem.org/Part:BBa_J364002 BBa_J364002]</li> | ||
| + | <li>Test Device 4 - [http://parts.igem.org/Part:BBa_J364003 BBa_J364003]</li> | ||
| + | <li>Test Device 5 - [http://parts.igem.org/Part:BBa_J364004 BBa_J364004]</li> | ||
| + | <li>Test Device 6 - [http://parts.igem.org/Part:BBa_J364005 BBa_J364005]</li> | ||
| + | </ol> | ||
| + | |||
<html> | <html> | ||
<br> | <br> | ||
<hr> | <hr> | ||
<br> | <br> | ||
| − | <h3> | + | <h3 style="font-family: verdana">Data</h3> |
| + | |||
</html> | </html> | ||
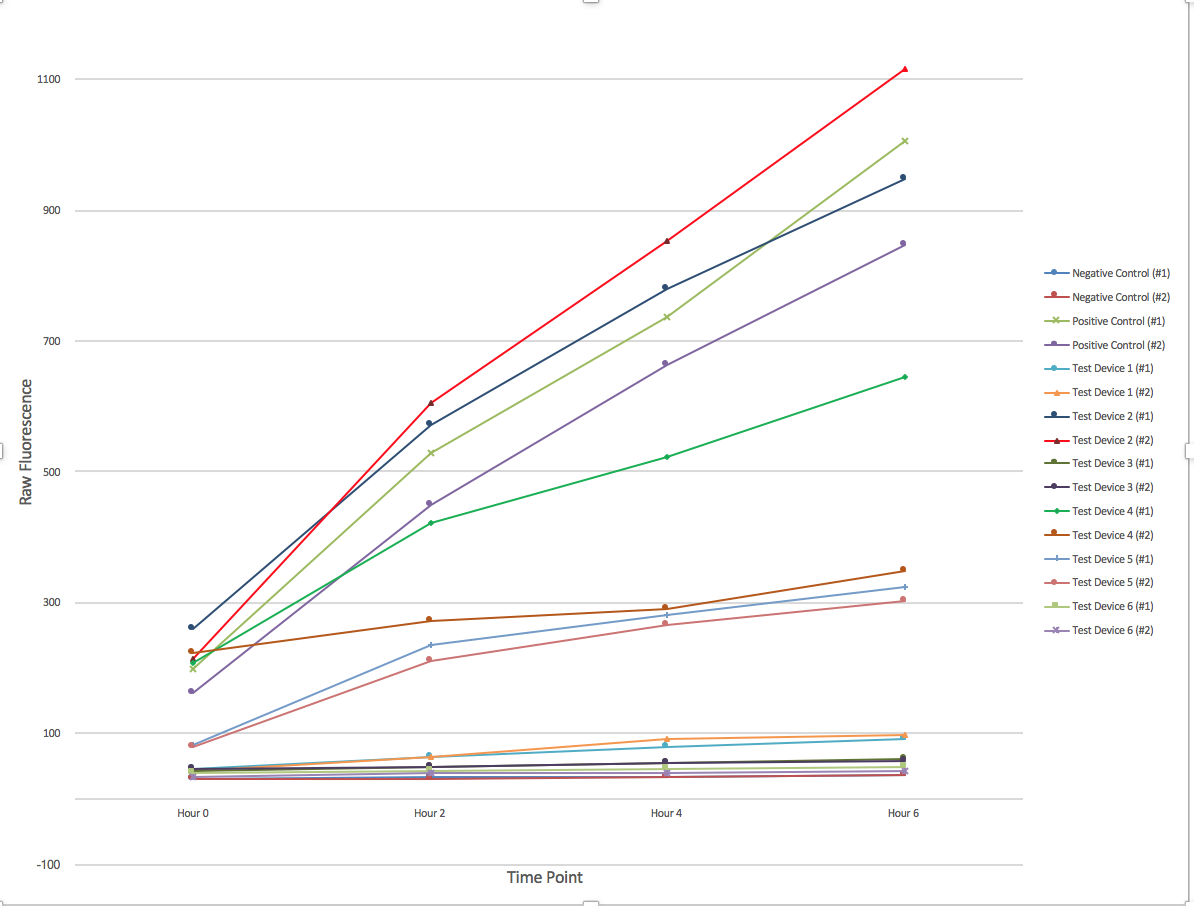
| − | + | [[File:T--Austin UTexas--interlabgraph.png|thumb|center|700px|Figure 1. The change of fluorescence over time. 6 different devices were employed in duplicate with a positive and negative control. Samples were collected every 2 hours for a total of 6 hours and afterwards had their fluorescence measured using a plate reader and 96-well plate. Graph produced by Ian Overman.]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<html> | <html> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</html> | </html> | ||
Latest revision as of 16:35, 1 November 2017
Interlab Measurement Study
The 2017 Interlab Study is the fourth such initiative by the Measurement Committee of iGEM Foundation to establish a hardy measurement procedure for the green fluorescent protein. The goal for the study this year is to test various RBS devices with the intention of finding which combinations yield more stable gene expression and how this might translate to various labs worldwide. Detailed below are the protocols and materials we used during the course of the experiment as provided by the iGEM Foundation. We are proud to contribute to this research endeavor by the iGEM Foundation.
The University of Texas at Austin iGEM team is one of nine teams to have participated in all of the interlab studies to date!
Protocol: Getting Started
- Transformed Interlab 2017 Part kit devices into E. coli DH5-alpha cells using electroporation.
- Plated on LB/CAM and allowed to grow overnight at 37° C.
- Two colonies from each plate used to inoculate 5 ml of LB/CAM liquid media and grown overnight at 37° C and 220 rpm for a total of sixteen culture tubes.
- Next dilutions were made in 12 ml of LB/CAM to a target OD600 of 0.02 and allowed to grow at 37° C and 220 rpm in a foil covered incubator with the lights off.
Protocol: Plate Reader
- Allow OD600 dilutions to grow for 6 hours at 37° C and 220 rpm
- Take measurements every two hours, starting with hour 0, for a total of four measurement points.
- Measurements were taken by:
- Taking 500µl from each of the sixteen culture tubes and put into microcentrifuge tubes.
- Next, 100µl from these samples were pipetted into the wells of a Costar black with clear-bottom 96 well plate.
- The samples were measured using an Infinite 200 Pro with the following settings: Excitation 485nm with a 9nm bandwidth and an Emission of 530nm with a bandwidth of 20nm. A gain of 48 was also used.
Ludox and Fluorescein Measurements
LUDOX-S40, provided by the iGEM foundation, was used to create a point of reference between the fluorescence data and OD600. Additionally, the provided fluorescein was used to create a standard curve of the fluorescence data gathered from the cellular measurements.
- LUDOX-S40 was measured by loading four, 100µl samples into a plate in wells A1, A2, A3, and A4, while 100µl diH2O was added to B1, B2, B3, and B4.
- Fluorescein was measured by diluting 2X fluorescein with PBS to 1X and brought to a total volume of 1 mL
- 100µl of PBS were loaded into the C1-F12 area on the same 96-well plate used for the LUDOX measurements.
- Serial dilutions were performed with 100µl of 1X fluorescein added to C1, mixed well, and then 100µl taken from that well and put into the next well, C2. This was continued to well C11, leaving C12 with just 100µl of PBS.
- The above procedure was repeated for rows D-F.
Devices Measured
All parts were located on the pSB1C3 plasmid backbone. The parts used as designated by the provided protocol were:
- Positive Control - [http://parts.igem.org/Part:BBa_I20270 BBa_I20270]
- Negative Control - [http://parts.igem.org/Part:BBa_R0040 BBa_R0040]
- Test Device 1 - [http://parts.igem.org/Part:BBa_J364000 BBa_J364000]
- Test Device 2 - [http://parts.igem.org/Part:BBa_J364001 BBa_J364001]
- Test Device 3 - [http://parts.igem.org/Part:BBa_J364002 BBa_J364002]
- Test Device 4 - [http://parts.igem.org/Part:BBa_J364003 BBa_J364003]
- Test Device 5 - [http://parts.igem.org/Part:BBa_J364004 BBa_J364004]
- Test Device 6 - [http://parts.igem.org/Part:BBa_J364005 BBa_J364005]
Data