| (103 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
{{Austin_UTexas}} | {{Austin_UTexas}} | ||
<html> | <html> | ||
| + | <style> | ||
| + | body { | ||
| + | background-color: #282828 | ||
| + | } | ||
| − | + | </style> | |
<div class="column full_size"> | <div class="column full_size"> | ||
| − | < | + | <div style="background-color: lightcyan;padding:14px; font-family: verdana"> |
| − | + | ||
| − | < | + | <h2 style="font-family: verdana">The Improved Blue Chromoprotein</h2> |
| − | < | + | <br> |
| + | <p style="font-family: verdana">Chromoproteins are known to be evolutionarily unstable, due to the fact that they create a high metabolic burden on the cell that they are in. This has been supported by studies conducted through various research. One experiment, done within this lab, shows the original blue chromoprotein, (<a href="http://parts.igem.org/Part:BBa_K592009">BBa_K592009</a></b>), streaked onto a plate where varying ranges of colored colonies grew, shown in <strong>Figure 1</strong>. Some of them appeared blue, while other did not, indicating the difference in the rate at which the blue chromoprotein breaks. This further supports the hypothesis that the original blue chromoprotein is unstable. Due to this instability, cells expressing the blue chromoprotein have a hard time retaining the blue phenotype. In order to combat this metabolic burden, a technique called codon optimization was used to improve the function of this part. Codon optimization involves exchanging certain codons for ones that have known to be more translationally efficient in a certain species while retaining the same sequence of amino acids. Making translation more efficient, in turn, lowers the metabolic burden in the cells that the blue chromoprotein gene is present in. | ||
| − | < | + | <p style="font-family: verdana">The codon optimized blue chromoprotein basic part can be found on the iGEM registry as: (<a href="http://parts.igem.org/Part:BBa_K2253002">BBa_K2253002</a></b>). |
| − | < | + | </p> |
| + | </html> | ||
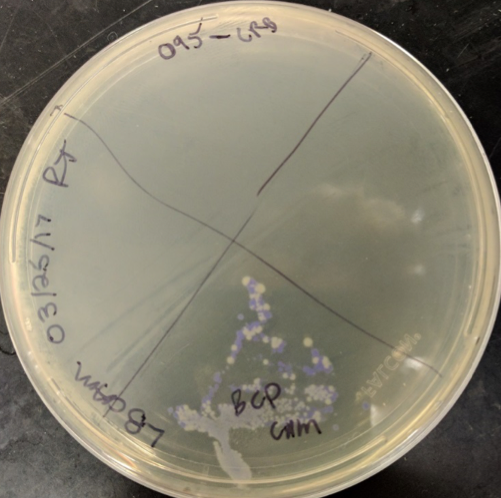
| + | [[File:Blue_cp_plate_background.png|thumb|center|400px|'''Figure 1.''' The image above shows various streaked out colonies from the original blue chromoprotein, BBa_K592009. Both phenotypically blue and clear colonies grew, supporting the hypothesis that the blue chromoprotein is unstable and breaks or changes color at different rates. Plate streaked by Microbe Hackers 2016 Chromoprotein Team. ]] | ||
| + | <html> | ||
| − | </ | + | <h4 style="font-family: verdana">Experimental Methods</h4> |
| + | <p style="font-family: verdana">The codon optimized sequence has been transformed into a plasmid with the pSB1C3 backbone (with the <a href="http://parts.igem.org/Part:BBa_K608002">BBa_K608002</a></b> promoter and RBS sequence) via electroporation. This transformation resulted in one phenotypically blue colony, shown in <b>Figure 2</b>. This colony was inoculated into liquid media to create an overnight culture, shown in <b>Figure 3</b>. The codon optimized sequence has been sequence verified and shows minimal point mutations in comparison to the original codon optimized blue chromoprotein sequence, derived from BBa_K592009. The sequence and amino acid sequence it codes for are shown in <strong>Figure 4.</strong> | ||
| + | <div class="column half_size" | ||
| + | </html> | ||
| + | |||
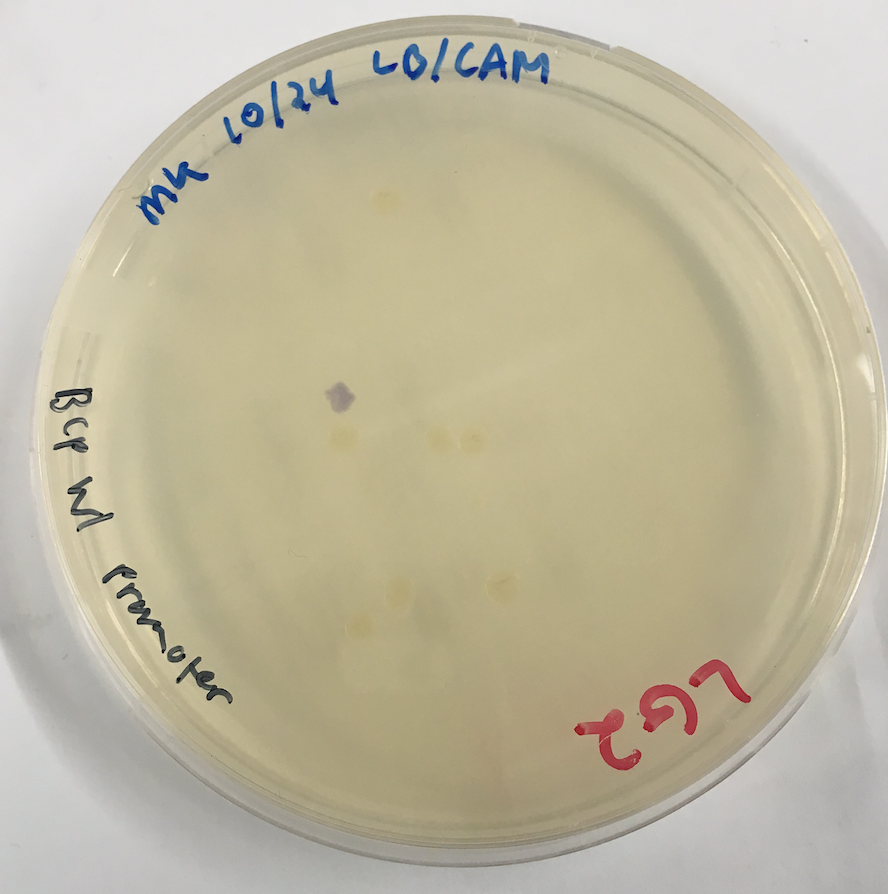
| + | [[File:T--Austin UTexas--blueplate.png|thumb|center|400px|'''Figure 2.''' The image above shows the transformation plate of the electroporation of the codon-optimized blue chromoprotein. The blue colony shown was used to make overnight cultures to miniprep and sequence.]] | ||
| + | <html> | ||
| + | </div> | ||
| + | <div class="column half_size" | ||
| + | </html> | ||
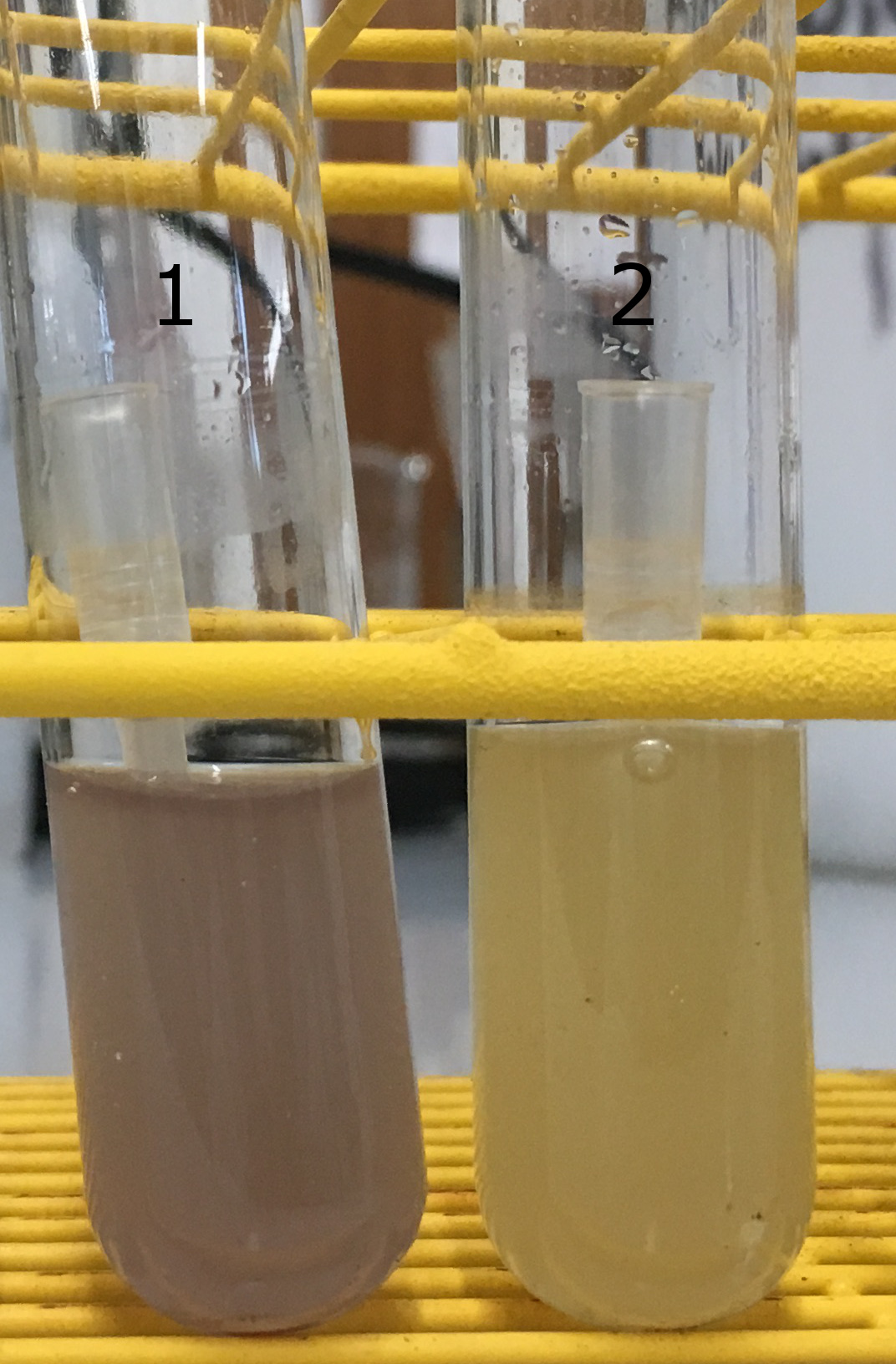
| + | [[File:T--Austin UTexas--BlueCPSide.jpg|thumb|center|250px|'''Figure 3.'''On the left is the overnight culture of the phenotypically blue colony from the codon-optimized blue chromoprotein transformation. On the right is normal ''E. coli'']] | ||
| + | <html> | ||
| + | </div> | ||
| + | |||
| + | </html> | ||
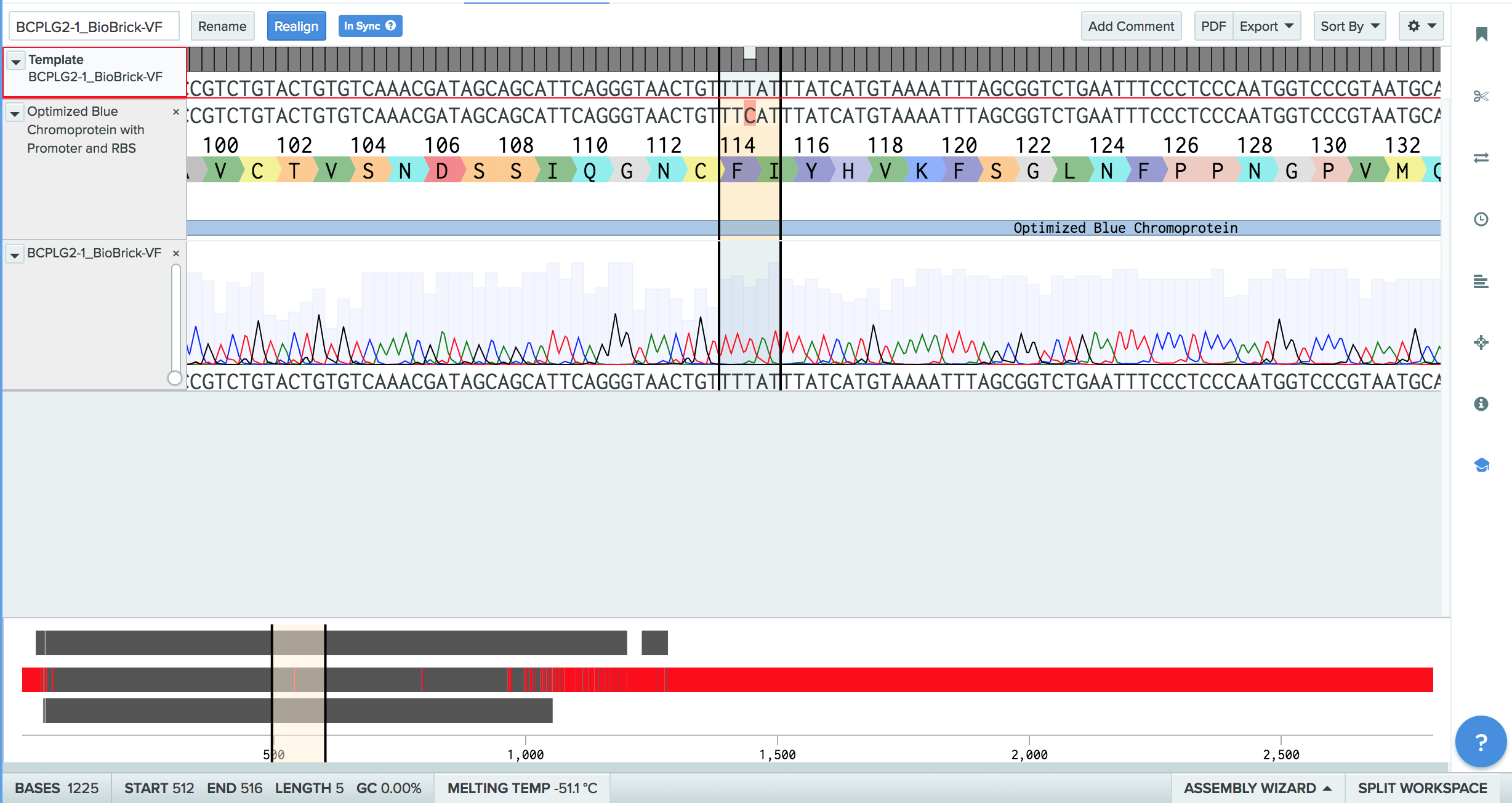
| + | [[File:Screen_Shot_2017-11-01_at_5.02.25_PM.png|thumb|center|800px|'''Figure 4.'''Benchling results show the point of the silent mutation, from 5’ TTT 3’ to 5’ TTC 3’ within the codon optimized sequence.'']] | ||
| + | <html> | ||
| + | |||
| + | <div class="clear"</div> | ||
| + | <h4 style="font-family: verdana">Improvements</h4> | ||
| + | <p style="font-family: verdana">Codon optimization greatly improves the function of the original blue chromoprotein (BBa_K592009) by making it more translationally efficient, and as stated earlier, lowers the metabolic burden on the <i>E. coli</i>. The lower metabolic burden, makes the E.coli less prone to mutations when the codon optimized sequence is inserted into the plasmid. The codon optimized sequence has been transformed into a plasmid with the pSB1C3 backbone with the K608002 promoter and RBS sequence and also has been sequence verified to be the correct amino acid sequence. There is one silent point mutation from 5’ TTT 3’ to 5’ TTC 3’ in the nucleotide sequence but it retains the amino acid phenylalanine and overall sequence as compared to the expected sequence of the codon optimized blue chromoprotein. Another improvement of this part is that the miniprepped sequence now has the promoter and RBS sequence attached to it, which can be taken and transformed immediately instead of having to go through the process of digestion and ligation all over again to add different parts to it. Prior to this, there was not an available blue chromoprotein sequence already ligated to the promoter and RBS sequence so this also improves the function of the blue chromoprotein part in it is ready to use without much additional work. | ||
| + | </p> | ||
| + | </div> | ||
</html> | </html> | ||
Latest revision as of 03:17, 2 November 2017
The Improved Blue Chromoprotein
Chromoproteins are known to be evolutionarily unstable, due to the fact that they create a high metabolic burden on the cell that they are in. This has been supported by studies conducted through various research. One experiment, done within this lab, shows the original blue chromoprotein, (BBa_K592009), streaked onto a plate where varying ranges of colored colonies grew, shown in Figure 1. Some of them appeared blue, while other did not, indicating the difference in the rate at which the blue chromoprotein breaks. This further supports the hypothesis that the original blue chromoprotein is unstable. Due to this instability, cells expressing the blue chromoprotein have a hard time retaining the blue phenotype. In order to combat this metabolic burden, a technique called codon optimization was used to improve the function of this part. Codon optimization involves exchanging certain codons for ones that have known to be more translationally efficient in a certain species while retaining the same sequence of amino acids. Making translation more efficient, in turn, lowers the metabolic burden in the cells that the blue chromoprotein gene is present in.
The codon optimized blue chromoprotein basic part can be found on the iGEM registry as: (BBa_K2253002).
Experimental Methods
The codon optimized sequence has been transformed into a plasmid with the pSB1C3 backbone (with the BBa_K608002 promoter and RBS sequence) via electroporation. This transformation resulted in one phenotypically blue colony, shown in Figure 2. This colony was inoculated into liquid media to create an overnight culture, shown in Figure 3. The codon optimized sequence has been sequence verified and shows minimal point mutations in comparison to the original codon optimized blue chromoprotein sequence, derived from BBa_K592009. The sequence and amino acid sequence it codes for are shown in Figure 4.
Improvements
Codon optimization greatly improves the function of the original blue chromoprotein (BBa_K592009) by making it more translationally efficient, and as stated earlier, lowers the metabolic burden on the E. coli. The lower metabolic burden, makes the E.coli less prone to mutations when the codon optimized sequence is inserted into the plasmid. The codon optimized sequence has been transformed into a plasmid with the pSB1C3 backbone with the K608002 promoter and RBS sequence and also has been sequence verified to be the correct amino acid sequence. There is one silent point mutation from 5’ TTT 3’ to 5’ TTC 3’ in the nucleotide sequence but it retains the amino acid phenylalanine and overall sequence as compared to the expected sequence of the codon optimized blue chromoprotein. Another improvement of this part is that the miniprepped sequence now has the promoter and RBS sequence attached to it, which can be taken and transformed immediately instead of having to go through the process of digestion and ligation all over again to add different parts to it. Prior to this, there was not an available blue chromoprotein sequence already ligated to the promoter and RBS sequence so this also improves the function of the blue chromoprotein part in it is ready to use without much additional work.