HK SKHLPSS (Talk | contribs) |
(→Peroxidase assay) |
||
| (25 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
{{HK_SKHLPSS_HEADER}} | {{HK_SKHLPSS_HEADER}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | = Material and Method = | |
| − | + | == Assemble of DNA Nano-cube == | |
| − | + | ||
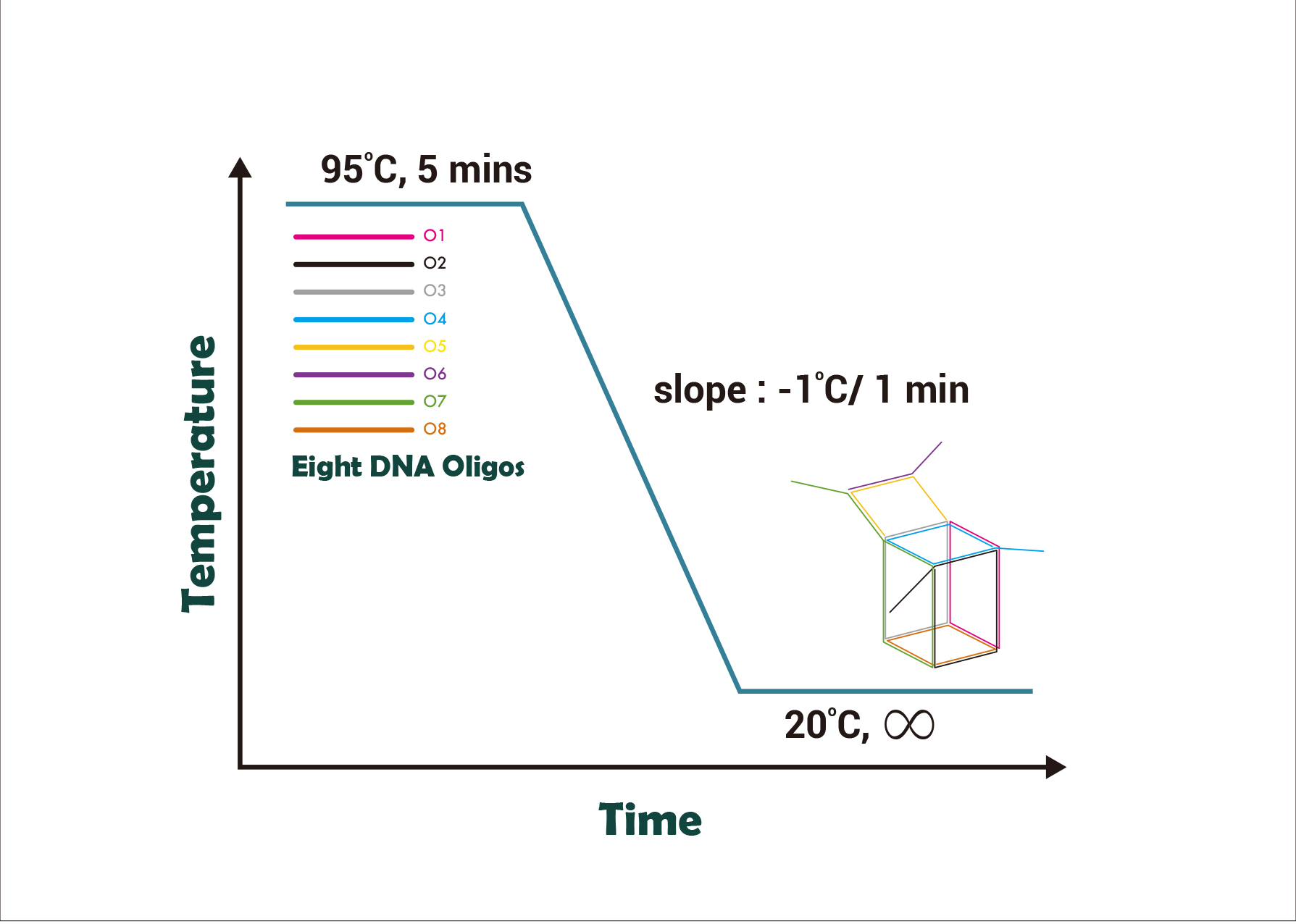
| − | + | Eight DNA strands are mixed and placed in the <I>Dynamica C-Master Gradient thermal cycler</I>. The thermal cycler raised the mixture’s temperature to 95°C for 5 minutes. The temperature was then maintained for 5 minutes. | |
| − | + | ||
| − | + | Then the temperature will be lowered 1°C per minute from 95°C to room temperature to allow eight single strands to anneal in accordance with our design by Watson & Crick’s base pairing to form the nano-cube. | |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[File:Material_and_Method_1.jpg |thumbnail| center | 500px| Fig 4. Annealing process of the nano-cube. The above figure shows that the whole annealing process involves eight oligos. The eight oligos will form a nano-cube when the temperature is lowered from 95°C to room temperature.]] | |
| − | + | ||
| − | + | ||
| − | + | == Native Polyacrylamide gel electrophoresis (Collaborated with HKU Team) == | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | Polyacrylamide gel electrophoresis (PAGE) is conducted to check whether the oligos are assembled as we designed. With assistance from iGEM HKU, the assembly of DNA nanostructure is analyzed by 12% PAGE where the combinations of oligos (10μL, 200nM) are loaded. All the gels are run at a constant voltage of 100V. GelRed is used to stain the gels. | ||
| − | + | For the analysis of target binding, equimolar (10μM final) DNA nanostructure and nucleic acid input are mixed and incubated at room temperature for 15 minutes in a shaker. The mixture (10μL, 200nM) is then loaded to 12% polyacrylamide gel. The PAGE is conducted at a constant voltage of 100V. GelRed is used to stain the gel. | |
| − | + | ||
| − | + | == Peroxidase assay == | |
| − | + | ||
| − | + | To prove the presence of the formed nano-cube, we conducted peroxidase assay by making use of a peroxidase activity of G-quadruplex. The target is RNA instead of DNA. However, to prove the concept, DNA peroxidase assay was done first because of its stability. RNA peroxidase assay was then followed. | |
| − | + | ||
| − | + | ||
| − | + | In our assay, nanocube (5 nM final), and hemin (10μM) are added to buffer. The ABTS and Hemin were purchased from Tin Hang Technology Ltd. The mixture was incubated at room temperature for 15 minutes in a <I>Dlab rocking shaker</I> at 60rpm. 15μL ABTS solution and H<sub>2</sub>O<sub>2</sub> (20mM final) are added to the mixture. The reaction mixture was then transferred to microplate and was read by <I>Dymanica Halo LED 96-well Microplate Reader</I> with an absorbance of 405 nm. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{HK_SKHLPSS_FOOTER}} | {{HK_SKHLPSS_FOOTER}} | ||
Latest revision as of 07:15, 13 December 2017
Material and Method
Assemble of DNA Nano-cube
Eight DNA strands are mixed and placed in the Dynamica C-Master Gradient thermal cycler. The thermal cycler raised the mixture’s temperature to 95°C for 5 minutes. The temperature was then maintained for 5 minutes.
Then the temperature will be lowered 1°C per minute from 95°C to room temperature to allow eight single strands to anneal in accordance with our design by Watson & Crick’s base pairing to form the nano-cube.
Native Polyacrylamide gel electrophoresis (Collaborated with HKU Team)
Polyacrylamide gel electrophoresis (PAGE) is conducted to check whether the oligos are assembled as we designed. With assistance from iGEM HKU, the assembly of DNA nanostructure is analyzed by 12% PAGE where the combinations of oligos (10μL, 200nM) are loaded. All the gels are run at a constant voltage of 100V. GelRed is used to stain the gels.
For the analysis of target binding, equimolar (10μM final) DNA nanostructure and nucleic acid input are mixed and incubated at room temperature for 15 minutes in a shaker. The mixture (10μL, 200nM) is then loaded to 12% polyacrylamide gel. The PAGE is conducted at a constant voltage of 100V. GelRed is used to stain the gel.
Peroxidase assay
To prove the presence of the formed nano-cube, we conducted peroxidase assay by making use of a peroxidase activity of G-quadruplex. The target is RNA instead of DNA. However, to prove the concept, DNA peroxidase assay was done first because of its stability. RNA peroxidase assay was then followed.
In our assay, nanocube (5 nM final), and hemin (10μM) are added to buffer. The ABTS and Hemin were purchased from Tin Hang Technology Ltd. The mixture was incubated at room temperature for 15 minutes in a Dlab rocking shaker at 60rpm. 15μL ABTS solution and H2O2 (20mM final) are added to the mixture. The reaction mixture was then transferred to microplate and was read by Dymanica Halo LED 96-well Microplate Reader with an absorbance of 405 nm.