| (34 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
{{:Team:SUSTech_Shenzhen/main-content-begin}} | {{:Team:SUSTech_Shenzhen/main-content-begin}} | ||
<!--------往下直接写内容-----------------> | <!--------往下直接写内容-----------------> | ||
| − | |||
| − | |||
| − | |||
| − | + | In our project, we want to study the neural network activity and behavioral response of Caenorhabditis elegans with orthogonal optogenetic stimulation of two neurons. | |
| − | + | So far, we have achieved almost all of our project objectives, building a platform to study the learning behavior of Caenorhabditis elegans. The whole project is demonstrated in the following parts: | |
| + | #Choosing the suitable excitation wavelength of light-sensitive proteins | ||
| + | #Genetic integration of synthetic circuits into C.elegans strains | ||
| + | #Examining the functions of AWA and AWB neurons | ||
| + | #Using Microfluidic device to examine the behavioral change of individual worms | ||
| + | #Training C.elegans to be addicted to alcohol | ||
| − | + | ------ | |
| − | + | ==Choosing the suitable excitation wavelength == | |
| − | + | Expression of two types of channelrhodopsins in two pairs of receptor neurons enables worm to be attracted by blue light and be repelled by red light. At the same time, we plan to use GEM-GECO as calcium indicator to examining the neuron activation, and perform feedback control in the future. | |
| − | + | ||
| − | + | To avoid potential crosstalk, we need to separate the excitation wavelengths of CoChR activation and GEM-GECO imaging. In our project, we chose 395nm and 470nm to activate CoChR and GEM-GECO. | |
| − | + | <html><!-----------------------------图?------------------------------></html> | |
| − | + | Before the experiment in worms, we conducted a validation experiment on CHO-K1 cell line expressing CoChR to identify a suitable excitation wavelength for the light-sensitive ion channel. | |
| − | {{SUSTech_Image_Center_fill-width| filename=T--SUSTech_Shenzhen-- | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig1.jpg|width=6000px|caption=<B> Fig.1 Results of experiment for CoChR spectrum determination.</B> In the experiment ,we expressed CoChR in the CHO-K1 cell line and R-GECO is used to detect the calcium influx into the cell. <B>A)</B> This figure shows that R-GECO in cells are activated after the 200mM KCl inducing. We take this group as the positive control group since we consider that the indicators were full-activated at this state. This is because 200mM KCl can cause plenty of calcium influx into the cell through calcium channel in cells membrane. <B>B)</B> This figure shows the light induced R-GECO activation, the light can cause the conformational change of CoChR, then cause calcium influx into the cell. The sharp peak on this curve proves that the light inducing is much more sensitive than the chemical signal. }} |
| − | + | Based on the result, we determine to use 470nm to activate the CoChR and 395nm for the excitation of GEM-GECO. | |
| − | + | ---------------- | |
| − | ---------- | + | ==Genetic integration of synthetic circuits into C.elegans strains== |
| − | == | + | |
| − | + | We used the miniMos system to integrate the gene circuits into C.elegans genome. After injection and selection (Fig.2), we successfully obtained the inserted worms (Odr-10::CoChR::GEM-GECO::mCherry and str-1::Chrimson::GEM-GECO::GFP) . | |
| − | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen-- | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig2.jpg|width=6000px|caption=<B>Fig.2 F1 Negative selection.</B> peel-1 is a negative selection marker used to select against animals carrying extra- chromosomal arrays and assist with identifying animals that have endogenous locus insertions. This marker was co-injected with the target gene along with the heatshock promoter, after heatshock the worms with multiple extra- chromosomal arrays will all die <B>(A)</B> while the worms with endogenous locus insertion will survive <B>(B)</B>. After 34°C,4h heatshock, these alive worms will be picked out for fluorescence checking.}} |
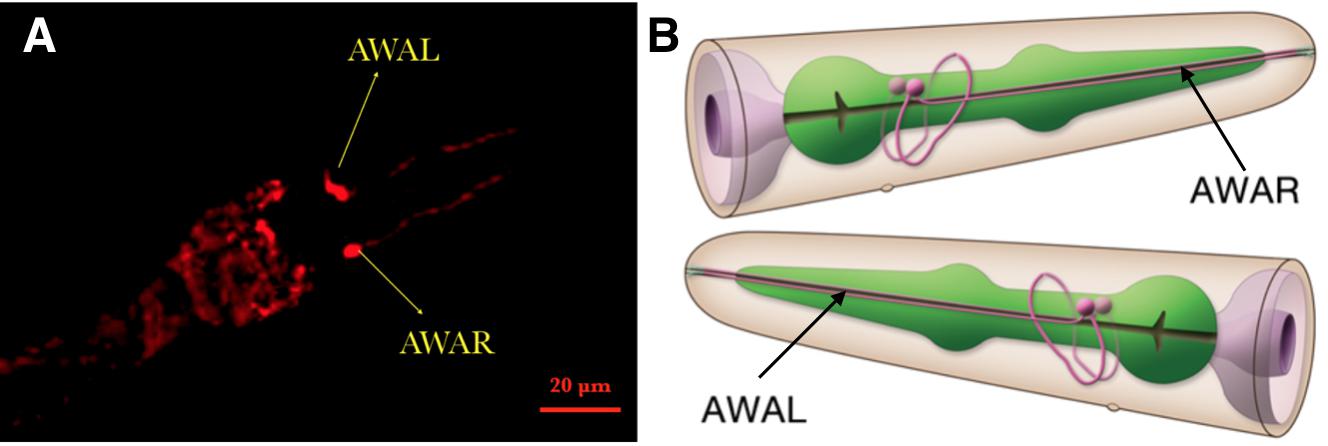
| − | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen-- | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig3.jpg|width=6000px|caption=<B>Fig.3 Confocal Image of Odr-10::CoCHR::GEM-GECO::mCherry worm’s head.</B> Through the red fluorescence of mCherry, we could determine the location of AWA neuron <B>(A)</B>. Compared to the 3D neuron model <B>(B)</B>, this result demonstrates that our circuit was successfully inserted and expressed in the C.elegans.}} |
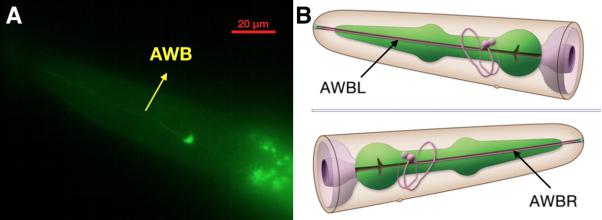
| − | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen-- | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig4.jpg|width=6000px|caption=<B>Fig.4 Confocal Image of str-1::Chrimson::GEM-GECO::GFP worm’s head.</B> Through the green fluorescence of GFP, we could determine the location of AWB neuron (A). Compared to the 3D neuron model <B>(B)</B>, this result demonstrates that our }} |
| − | + | ||
| − | + | ||
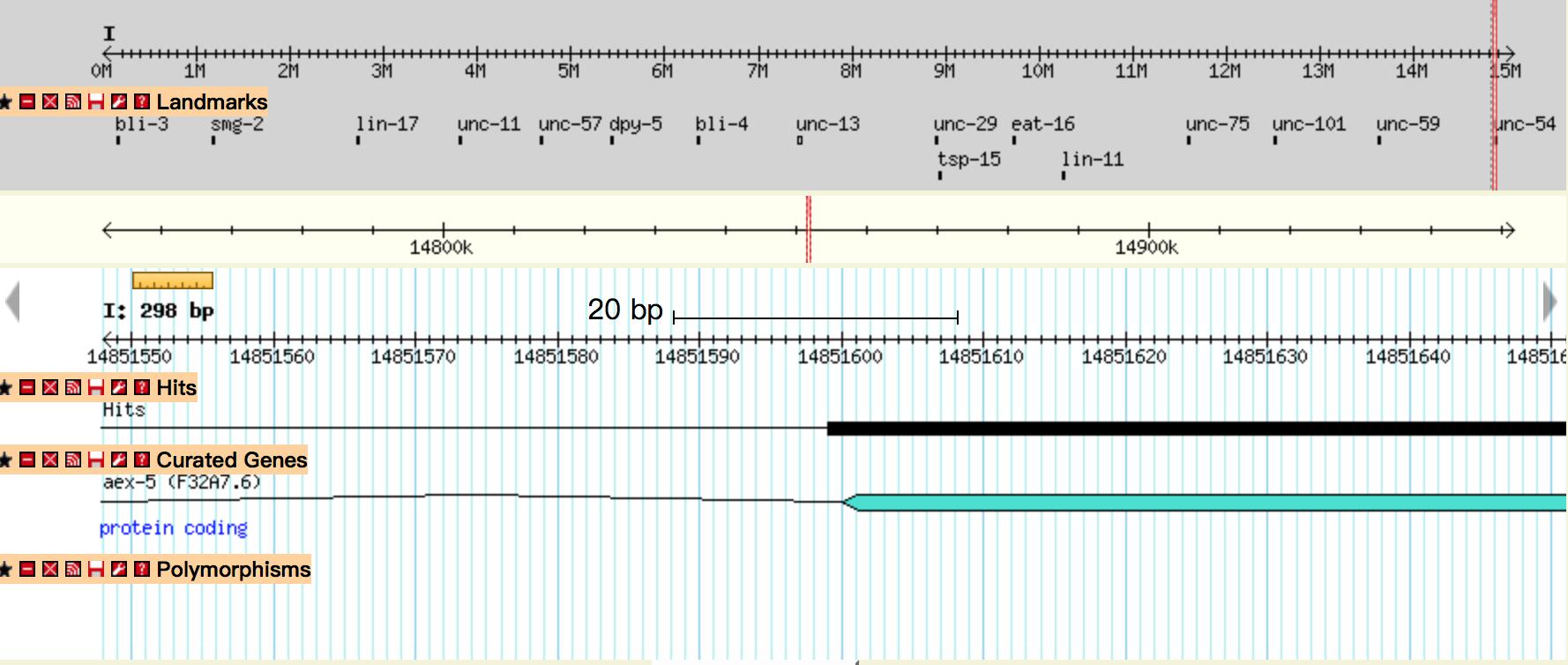
| − | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig5.jpg|width=6000px|caption=<B>Fig 5.</B> The mapping results of circuit Odr-10::CoCHR::GEM-GECO::mCherry after BLAST on wormbase.org. The black line represents location of the gene we insert. The comparison result showed that our gene inserted in the chromosome I and started at 14851599. }} | |
| − | + | After getting the genetically inserted worms, we successfully got the hybrid by mating 2 strains. | |
| − | == | + | ==Examining the function of AWA and AWB neurons== |
| − | + | Firstly, the experiment in which inserted worms are strongly attracted by diacetyl shows that the insertion did not degrade normal neuron function (Fig.6) | |
| − | + | We also test the result in the neuron level (Fig.7) | |
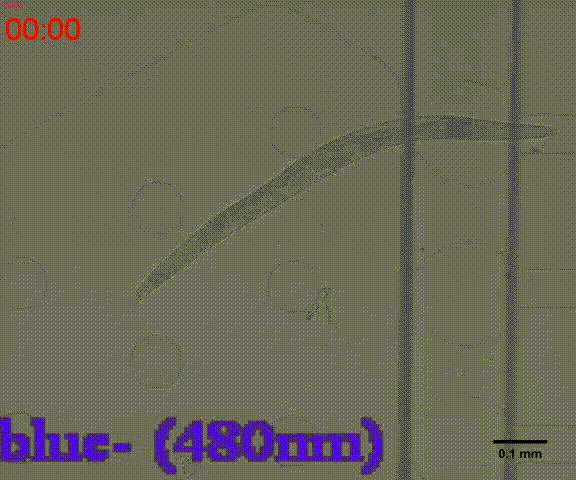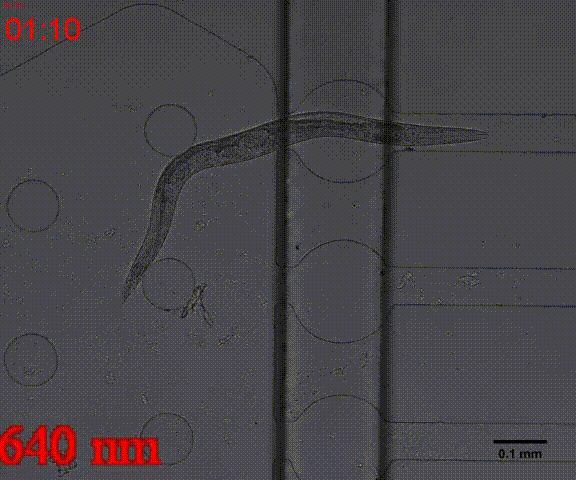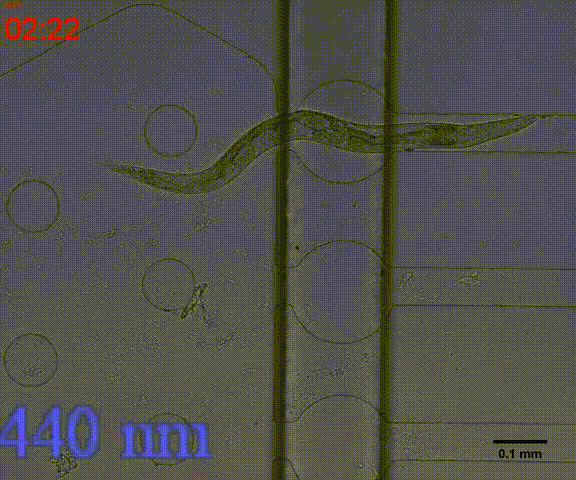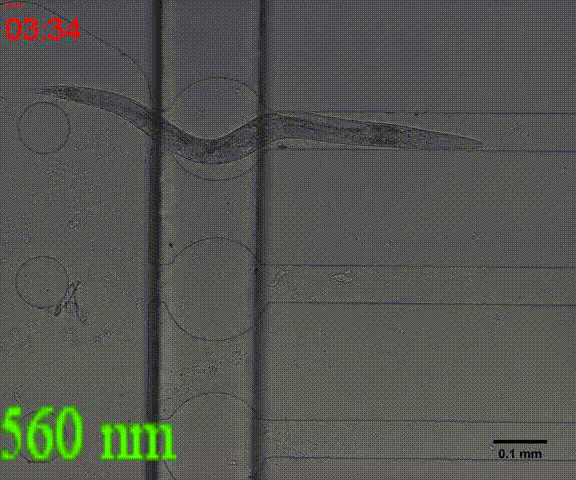
| − | {{SUSTech_Image_Center_8 | filename=T--SUSTech_Shenzhen-- | + | {{SUSTech_Image_Center_8 | filename=T--SUSTech_Shenzhen--3.gif|width=6000px|caption=<B>Fig. 6 C.elegans' locomotion in the Gaussian plate</B> C.elegans can move forward freely in the channel, and then we will get its locomotion distribution just like the Galton Nail Plate model. However, after adding diacetyl in the side channel, the distribution is changed. C.elegans tend to move towards the right (or the left) due to their preference to diacetyl.}} |
| − | + | ||
| − | + | ||
| − | + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig7.png|width=6000px|caption=<B>Fig.7 Demonstrate the function of AWA neuron A)</B> Confocal Image of worm’s head neuron cells of AWA neurons are circled at the photo (yellow) <B>B)</B> The fluorescence intensity of 497-527nm emission of GEM-GECO falls steeply after applying diacetyl. Combined with calcium, the emission wavelength of GEM-GECO shift from 510nm to 470nm ( central peak)}} | |
| − | |||
| − | + | ==Examining the behavioral change of individual worms == | |
| + | |||
| + | We designed two methods to observe inserted worms’ behavior change: | ||
| + | #Inducing free-moving worms by certain wavelength (Fig.8A) | ||
| + | #Stimulating worms semi-fixed in microfluidic chips (Fig.8B) | ||
| + | |||
| + | In the inducing experiments, Odr-10::CoCHR::GEM-GECO::mCherry worms shows a strong preference to blue light. In the later experiment, we will use this preference to train them to form specific behavior. | ||
| + | |||
| + | Unfortunately, str-1::Chrimson::GEM-GECO::GFP worms are not clearly repelled by the red light. The low sensitivity is probably due to the expression level of Chrimson under the control of str-1 promoter. An modified circuit containing amplification step is design, but the implementation cannot meet the deadline of this wiki. | ||
<html> | <html> | ||
| − | <div class="col-md- | + | <div class="row"> |
| − | </ | + | <div class="col-md-1"></div> |
| − | + | <div class="col-md-10"> | |
| − | < | + | <video width="600px" controls="controls" autoplay="autoplay"> |
| − | <div class="col-md- | + | <source src="https://static.igem.org/mediawiki/2017/9/9f/T--SUSTech_Shenzhen--Microfuildics--tracking.mp4" type="video/ogg" /> |
| + | </video> | ||
| + | <p ><B>Fig. 8A Behavior experiments of C.elegans.</B> These two figures show the worm have obvious response to the blue light which could activate channelrhodopsin CoChR. the first video shows the worm which expressed Odr-10::CoChR::GEM-GECO::mCherry circuit could run in circle to follow the blue light. The other figure shows the same type worm which looks “asleep” ( because of no food supply) be “waked up” also by blue light(preference). </p> | ||
| + | |||
| + | </div> | ||
| + | <div class="col-md-1"></div> | ||
| + | </div> | ||
| + | |||
| + | <div class="row"> | ||
</html> | </html> | ||
| − | {{ | + | |
| + | {{SUSTech_Image_Center_8 | filename=T--SUSTech_Shenzhen--Microfuildics--wake up.gif|width=1000px|caption=<B>Fig 8B. Observe individual worms in microfluidic chip</B>. This PDMS channel is designed to fix worms and observe them under stereoscope. 4 channels with decreasing diameter is used to fix worm. Other 4 narrow channels allow worms to move in a line constraint. }} | ||
<html></div></br></br></br></html> | <html></div></br></br></br></html> | ||
| − | ==Alcohol addiction== | + | ==Alcohol addiction == |
| + | |||
| + | We trained the worms engineered with Odr-10::CoChR::GEM-GECO::mCherry to be “addicted to” alcohol successfully. | ||
| + | #In this experiment, we added the alcohol on the NGM plate containing engineered worms. At the same time, blue light is turned on to stimulate worms continually. | ||
| + | #After training for 2 hours, we washed the plate and recovered the worms in M9 buffer. | ||
| + | #Then, we put the mixture contained worms and M9 buffer on one side of a new plate. After recovery of the worms, we added the alcohol on the other side of the same plate to see if the worms have the tropism to the alcohol. | ||
| + | |||
| + | The result shows that most of the worms transfected with Odr-10::CoChR::GEM-GECO::mCherry moved to the alcohol while the wild types could not (Fig.9). | ||
| + | |||
| + | What’s more, without the previous training by light, the worms transfected with Odr-10::CoChR::GEM-GECO::mCherry show no preference to alcohol, neither. (Fig.10). | ||
| + | |||
| + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Demonstrate-fig9.10.jpg|width=6000px|caption=<B>Fig. 9 A.)</B> The behaviors of Odr-10::CoChR::GEM-GECO::mCherry worms (all stages) after 2 hours' training. We circle the area of worms after training with blue light in red. <B>B)</B> The same worms in figure A would crawl towards alcohol inducing. The yellow circle area represent the alcohol. <B>C)</B> The wild type worms control. <B>D)</B> The wild types would not crawl towards the alcohol even thought training with blue light. | ||
| + | <B>Fig. 10 E)</B> The buffer have no addition to worms transfected with Odr-10::CoChR::GEM-GECO::mCherry circuit. <B>F)</B> shows no preference to alcohol }} | ||
| + | |||
| + | This experiment showed that our genetically modified worms can learn new behavior(addiction to alcohol) after trained by blue light. | ||
| + | |||
| + | {|class="table table-striped" | ||
| + | |- | ||
| + | | Type of C. elegans | ||
| + | | Total Number of the Worms | ||
| + | | The number of the Induced Worms | ||
| + | | Ratio (Induced worms/Total worms) | ||
| + | |- | ||
| + | | Odr10::CoChR::GEM-GECO::mCherry worms-training-alcohol induce | ||
| + | | 1280 | ||
| + | | 154 | ||
| + | | 12.03% | ||
| + | |- | ||
| + | | Odr10::CoChR::GEM-GECO::mCherry worms -training-buffer induce | ||
| + | | 320 | ||
| + | | 6 | ||
| + | | 1.88% | ||
| + | |- | ||
| + | | Odr10::CoChR::GEM-GECO::mCherry worms -notraining-alcohol induce | ||
| + | | 256 | ||
| + | | 2 | ||
| + | | 0.78% | ||
| + | |- | ||
| + | | wild types-training-alcohol induce | ||
| + | | 220 | ||
| + | | 1 | ||
| + | | 0.45% | ||
| + | |} | ||
| + | |||
| + | ==Future Plan== | ||
| + | |||
| + | Our further work is to decipher the change of neuron connectivity due to the newly learned behavior. | ||
| + | |||
| + | We have selected 2 promoters(cho-1 and unc-8) to express optogenetic circuits on the neurons connected to AWA and AWB, such as AIA, AIY, VB, ASH, AVB, VB, AVA.<html><a href="https://2017.igem.org/Team:SUSTech_Shenzhen/Neuron_Network_Model">See details</a></html> The next step will be using optofluidics platform above to analyze the activation of downstream neurons in the new-formed or modulated neural circuit, with or without the aforementioned training. | ||
Latest revision as of 01:36, 15 December 2017
Demonstrate
Project
Contents
In our project, we want to study the neural network activity and behavioral response of Caenorhabditis elegans with orthogonal optogenetic stimulation of two neurons.
So far, we have achieved almost all of our project objectives, building a platform to study the learning behavior of Caenorhabditis elegans. The whole project is demonstrated in the following parts:
- Choosing the suitable excitation wavelength of light-sensitive proteins
- Genetic integration of synthetic circuits into C.elegans strains
- Examining the functions of AWA and AWB neurons
- Using Microfluidic device to examine the behavioral change of individual worms
- Training C.elegans to be addicted to alcohol
Choosing the suitable excitation wavelength
Expression of two types of channelrhodopsins in two pairs of receptor neurons enables worm to be attracted by blue light and be repelled by red light. At the same time, we plan to use GEM-GECO as calcium indicator to examining the neuron activation, and perform feedback control in the future.
To avoid potential crosstalk, we need to separate the excitation wavelengths of CoChR activation and GEM-GECO imaging. In our project, we chose 395nm and 470nm to activate CoChR and GEM-GECO.
Before the experiment in worms, we conducted a validation experiment on CHO-K1 cell line expressing CoChR to identify a suitable excitation wavelength for the light-sensitive ion channel.
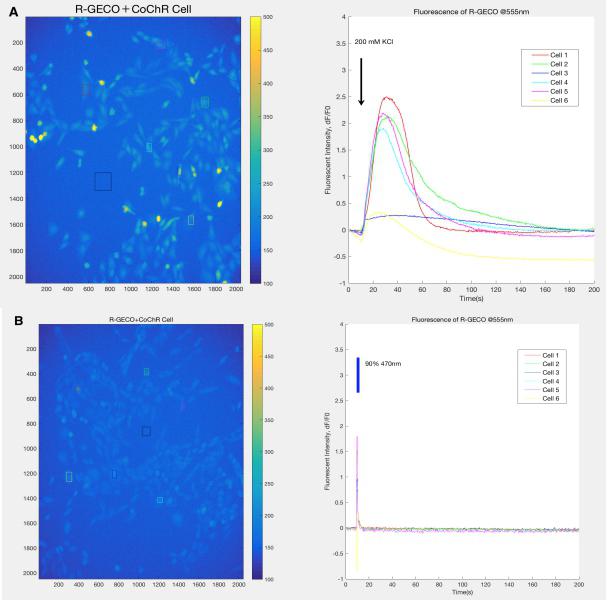
Based on the result, we determine to use 470nm to activate the CoChR and 395nm for the excitation of GEM-GECO.
Genetic integration of synthetic circuits into C.elegans strains
We used the miniMos system to integrate the gene circuits into C.elegans genome. After injection and selection (Fig.2), we successfully obtained the inserted worms (Odr-10::CoChR::GEM-GECO::mCherry and str-1::Chrimson::GEM-GECO::GFP) .
After getting the genetically inserted worms, we successfully got the hybrid by mating 2 strains.
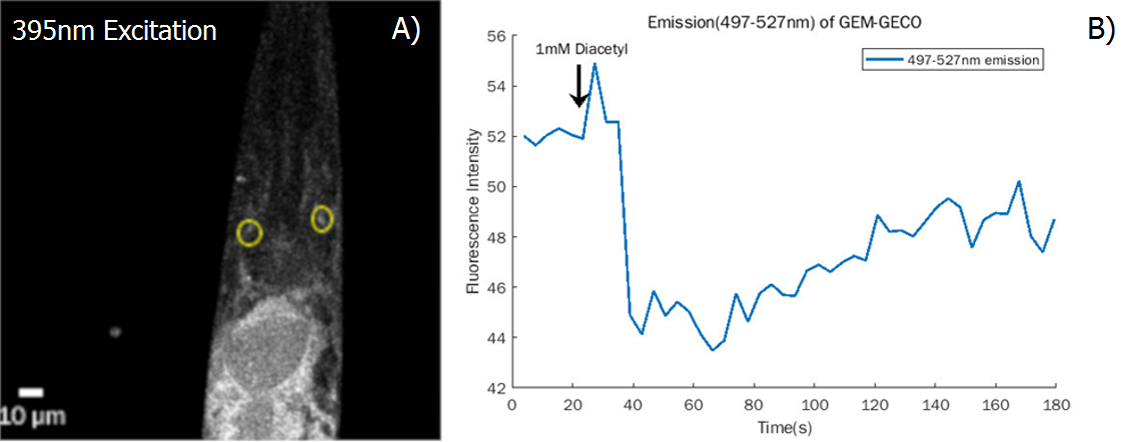
Examining the function of AWA and AWB neurons
Firstly, the experiment in which inserted worms are strongly attracted by diacetyl shows that the insertion did not degrade normal neuron function (Fig.6)
We also test the result in the neuron level (Fig.7)

Examining the behavioral change of individual worms
We designed two methods to observe inserted worms’ behavior change:
- Inducing free-moving worms by certain wavelength (Fig.8A)
- Stimulating worms semi-fixed in microfluidic chips (Fig.8B)
In the inducing experiments, Odr-10::CoCHR::GEM-GECO::mCherry worms shows a strong preference to blue light. In the later experiment, we will use this preference to train them to form specific behavior.
Unfortunately, str-1::Chrimson::GEM-GECO::GFP worms are not clearly repelled by the red light. The low sensitivity is probably due to the expression level of Chrimson under the control of str-1 promoter. An modified circuit containing amplification step is design, but the implementation cannot meet the deadline of this wiki.
Fig. 8A Behavior experiments of C.elegans. These two figures show the worm have obvious response to the blue light which could activate channelrhodopsin CoChR. the first video shows the worm which expressed Odr-10::CoChR::GEM-GECO::mCherry circuit could run in circle to follow the blue light. The other figure shows the same type worm which looks “asleep” ( because of no food supply) be “waked up” also by blue light(preference).
Alcohol addiction
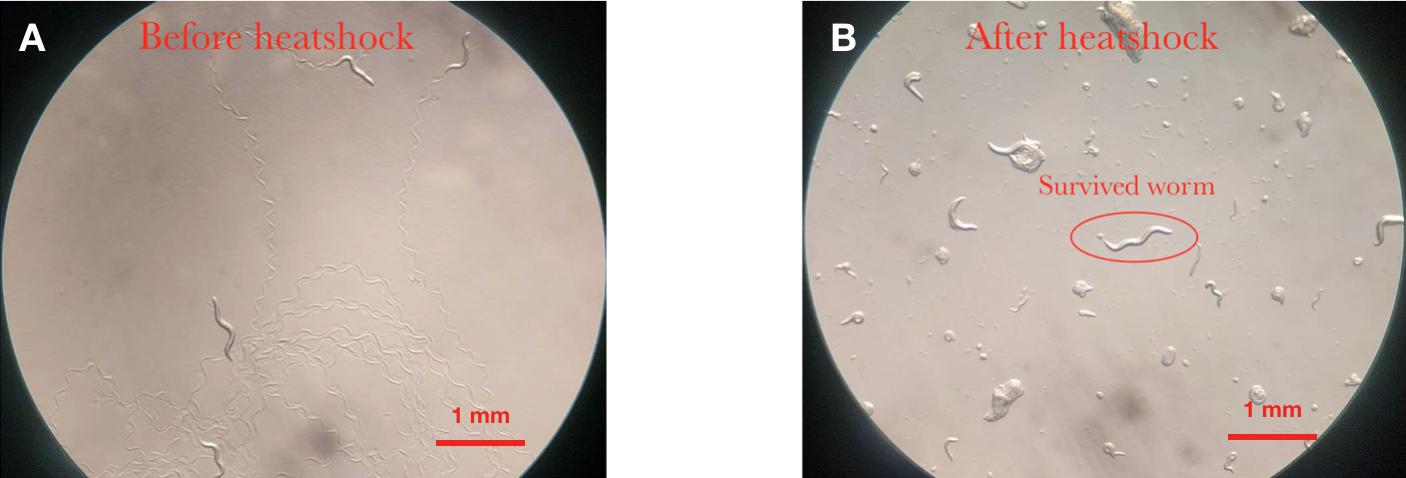
We trained the worms engineered with Odr-10::CoChR::GEM-GECO::mCherry to be “addicted to” alcohol successfully.
- In this experiment, we added the alcohol on the NGM plate containing engineered worms. At the same time, blue light is turned on to stimulate worms continually.
- After training for 2 hours, we washed the plate and recovered the worms in M9 buffer.
- Then, we put the mixture contained worms and M9 buffer on one side of a new plate. After recovery of the worms, we added the alcohol on the other side of the same plate to see if the worms have the tropism to the alcohol.
The result shows that most of the worms transfected with Odr-10::CoChR::GEM-GECO::mCherry moved to the alcohol while the wild types could not (Fig.9).
What’s more, without the previous training by light, the worms transfected with Odr-10::CoChR::GEM-GECO::mCherry show no preference to alcohol, neither. (Fig.10).
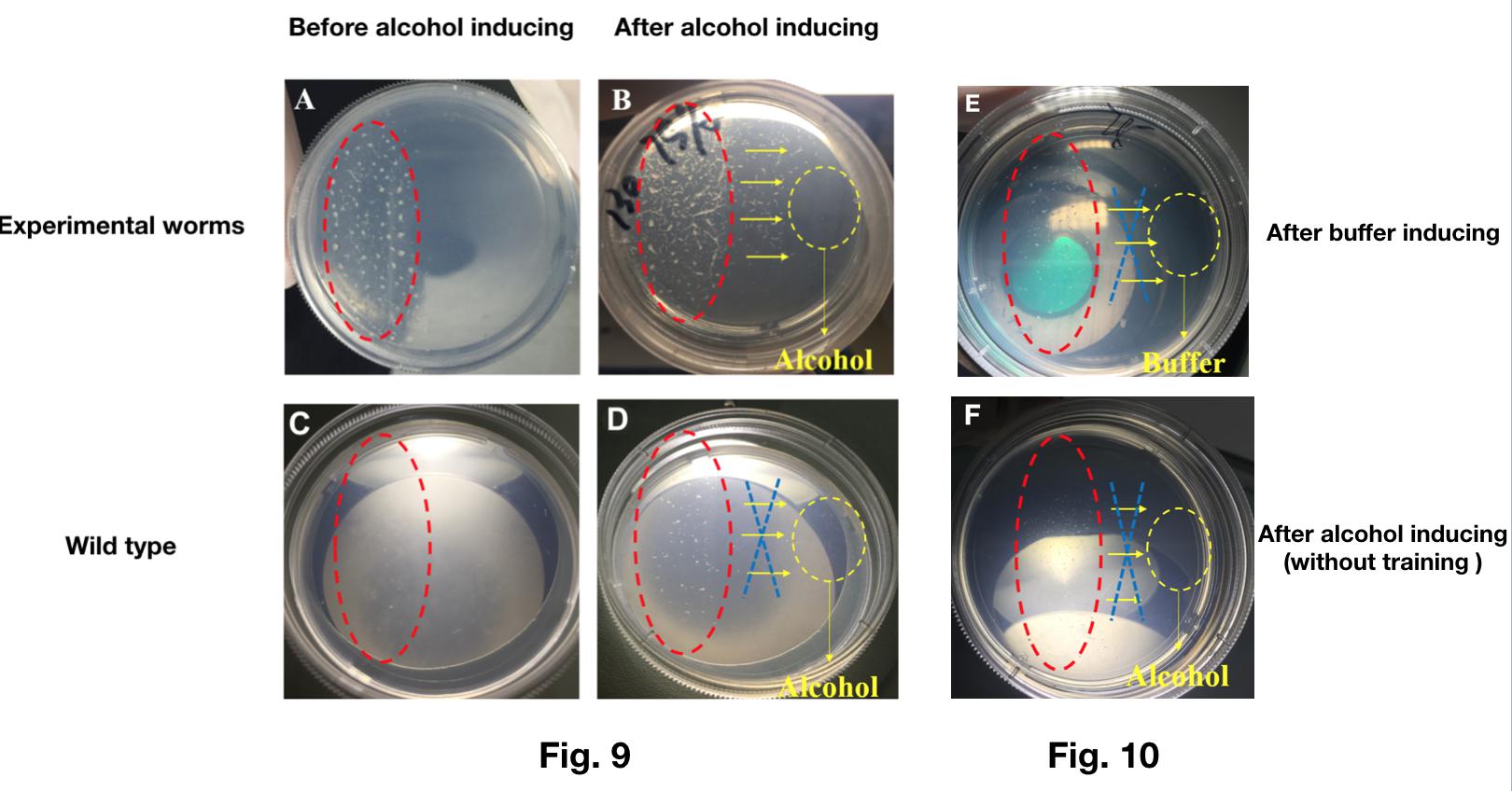
This experiment showed that our genetically modified worms can learn new behavior(addiction to alcohol) after trained by blue light.
| Type of C. elegans | Total Number of the Worms | The number of the Induced Worms | Ratio (Induced worms/Total worms) |
| Odr10::CoChR::GEM-GECO::mCherry worms-training-alcohol induce | 1280 | 154 | 12.03% |
| Odr10::CoChR::GEM-GECO::mCherry worms -training-buffer induce | 320 | 6 | 1.88% |
| Odr10::CoChR::GEM-GECO::mCherry worms -notraining-alcohol induce | 256 | 2 | 0.78% |
| wild types-training-alcohol induce | 220 | 1 | 0.45% |
Future Plan
Our further work is to decipher the change of neuron connectivity due to the newly learned behavior.
We have selected 2 promoters(cho-1 and unc-8) to express optogenetic circuits on the neurons connected to AWA and AWB, such as AIA, AIY, VB, ASH, AVB, VB, AVA.See details The next step will be using optofluidics platform above to analyze the activation of downstream neurons in the new-formed or modulated neural circuit, with or without the aforementioned training.