| Line 116: | Line 116: | ||
</div> | </div> | ||
| + | <div class="naviSection" id="section2"> | ||
| + | <br><br><br> | ||
<h2 style="font-family: verdana; font-size: 35px">Testing constitutive <i>Lactoccal</i> promoters </h2> | <h2 style="font-family: verdana; font-size: 35px">Testing constitutive <i>Lactoccal</i> promoters </h2> | ||
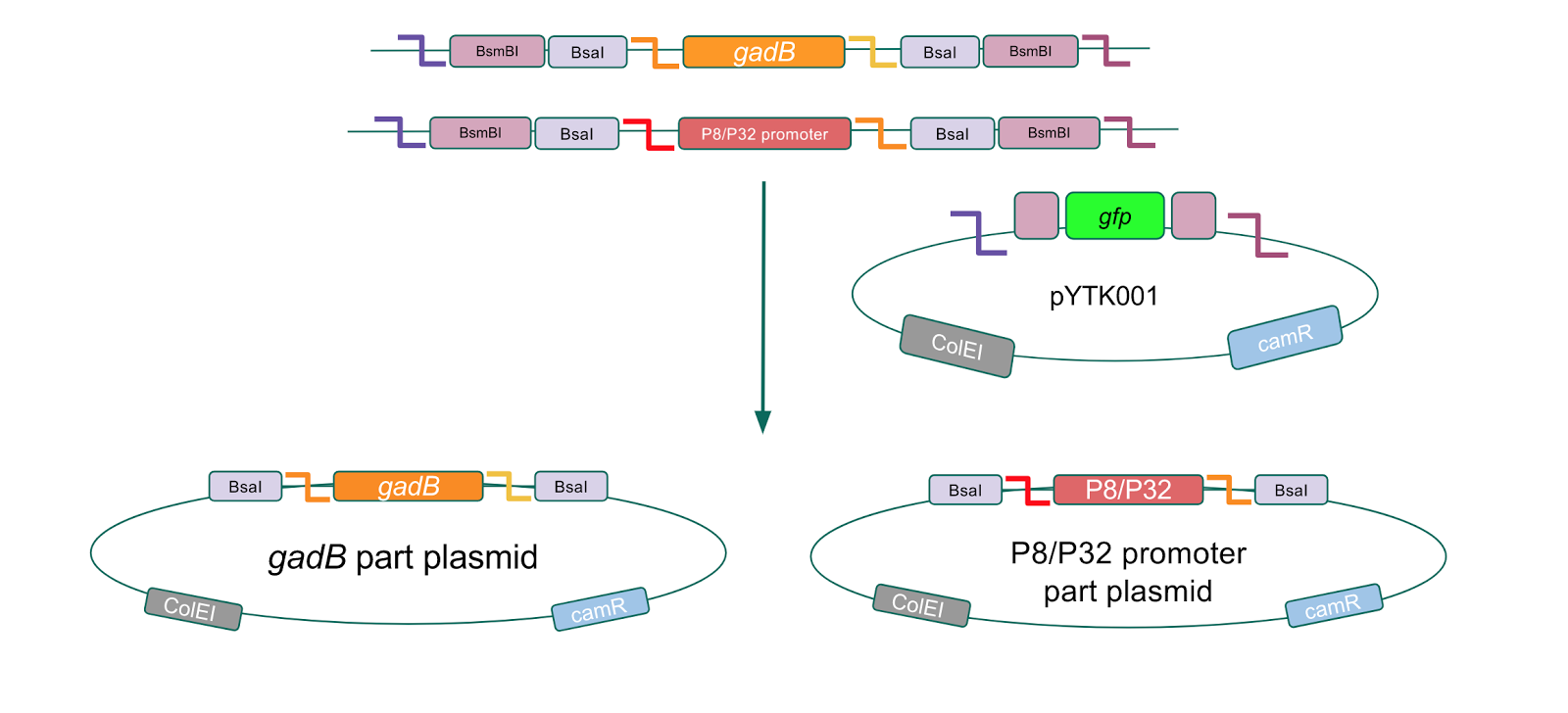
<p style="font-family: verdana">After successfully creating the <i>gadB</i> gene and P8/P32 promoter part plasmids, the functionality of these part plasmids were then assessed by assembling them into cassette plasmids.</p> | <p style="font-family: verdana">After successfully creating the <i>gadB</i> gene and P8/P32 promoter part plasmids, the functionality of these part plasmids were then assessed by assembling them into cassette plasmids.</p> | ||
| Line 145: | Line 147: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | + | <a href="#top"><p>Back to Top</p></a> | |
| + | </div> | ||
<h2 style="font-family: verdana; font-size: 35px">Testing for <i>gadB</i> overexpression in <i>E. coli</i></h2> | <h2 style="font-family: verdana; font-size: 35px">Testing for <i>gadB</i> overexpression in <i>E. coli</i></h2> | ||
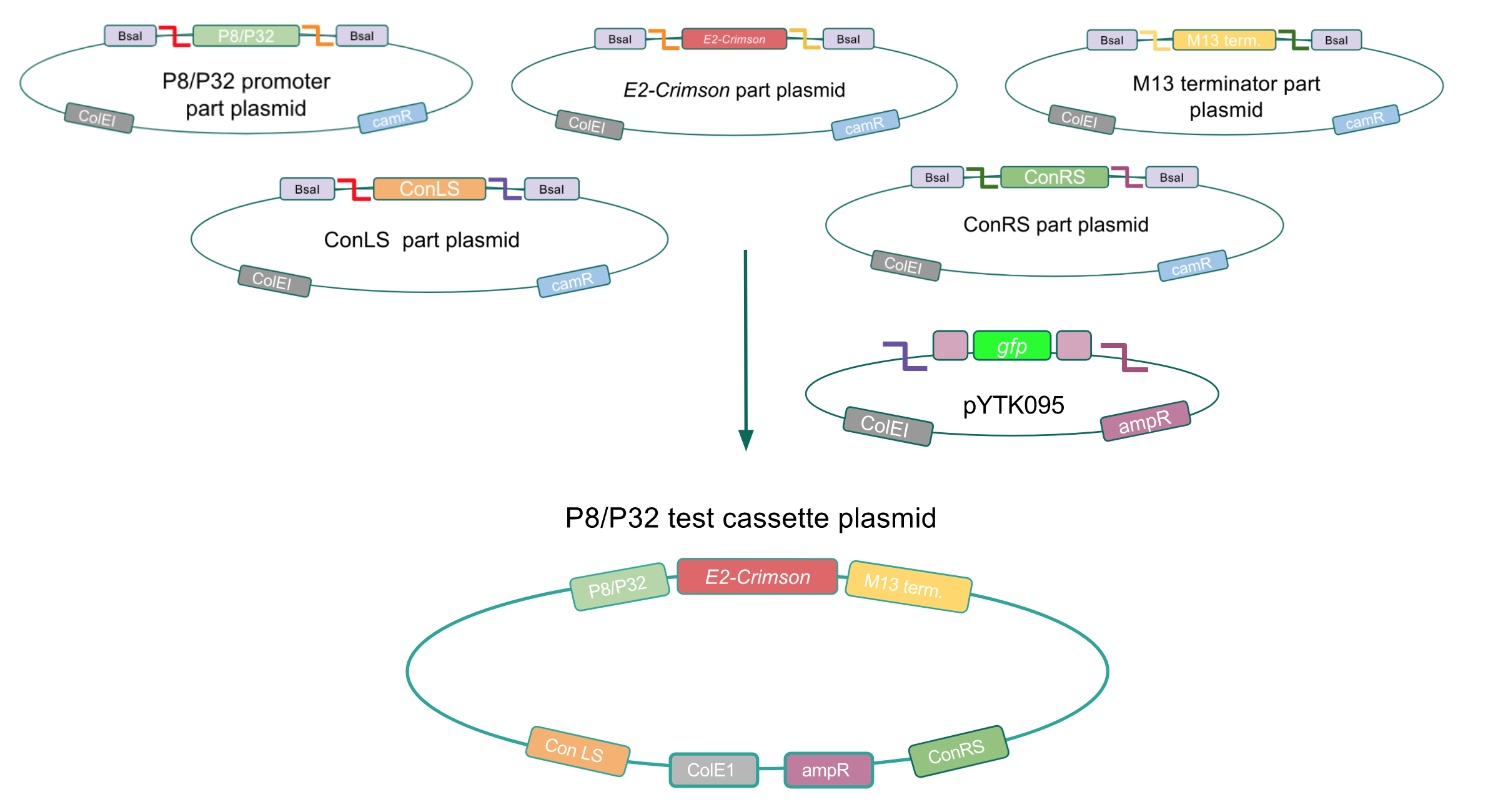
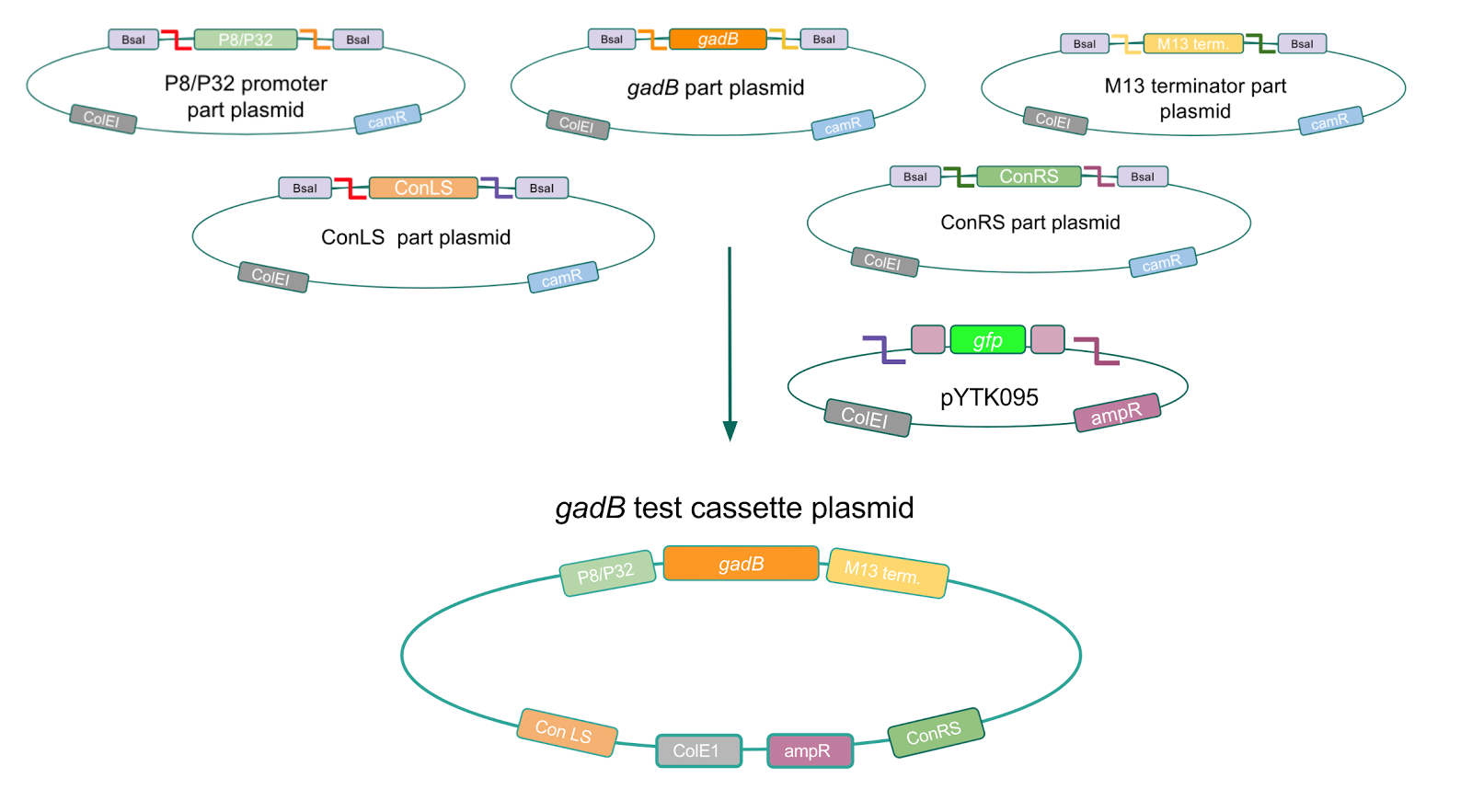
<p style="font-family: verdana">Using Golden Gate Assembly, we created cassette plasmids to test if the <i>gadB</i> gene could be overexpressed in <i>E. coli</i> via the P8 and P32 promoters. These cassette plasmids contained the <i>gadB</i> gene inserted downstream of either the P8 or P32 promoters. To make our <i>gadB</i> test cassette plasmids, we used pYTK095 as our backbone and part plasmids containing the P8/P32 promoters, <i>gadB</i> gene, M13 terminator, and connector sequences (Fig. 5).</p> | <p style="font-family: verdana">Using Golden Gate Assembly, we created cassette plasmids to test if the <i>gadB</i> gene could be overexpressed in <i>E. coli</i> via the P8 and P32 promoters. These cassette plasmids contained the <i>gadB</i> gene inserted downstream of either the P8 or P32 promoters. To make our <i>gadB</i> test cassette plasmids, we used pYTK095 as our backbone and part plasmids containing the P8/P32 promoters, <i>gadB</i> gene, M13 terminator, and connector sequences (Fig. 5).</p> | ||
Revision as of 23:08, 31 October 2017
Click on one of the images below to learn more about our results!
Golden Gate Assembly
Although bacteria can naturally synthesize GABA, we wanted to increase expression of the gadB gene and subsequently GABA production in order to imbue a more potent medicinal quality to our probiotic, with the idea that this GABA-overproducing probiotic can then be consumed by patients with bowel disorders or anxiety (1). Overexpression of the gadB gene will be accomplished by placing it under the control of either the P8 or P32 constitutive promoters from Lactococcus lactis (2).
To make our GABA-producing probiotic we first needed to assemble a GABA overexpression cassette plasmid using the Golden Gate assembly method. The intention here is that bacteria containing this GABA overexpression cassette plasmid should produce high levels of GABA. In short, Golden Gate Assembly is a new cloning method that allows for the creation of a multi-part DNA assembly (i.e. cassette plasmid) in a single reaction through the use of DNA parts containing specific, predefined suffixes and prefixes with recognition sites for Type IIs restriction enzymes (e.g. BsmBI and BsaI). The specificity of these suffixes and prefixes provides directionality of the desired DNA parts during the assembly process. For our purposes, we used the MoClo Yeast Tool Kit by John Dueber (3).
Testing for gadB overexpression in E. coli
Using Golden Gate Assembly, we created cassette plasmids to test if the gadB gene could be overexpressed in E. coli via the P8 and P32 promoters. These cassette plasmids contained the gadB gene inserted downstream of either the P8 or P32 promoters. To make our gadB test cassette plasmids, we used pYTK095 as our backbone and part plasmids containing the P8/P32 promoters, gadB gene, M13 terminator, and connector sequences (Fig. 5).
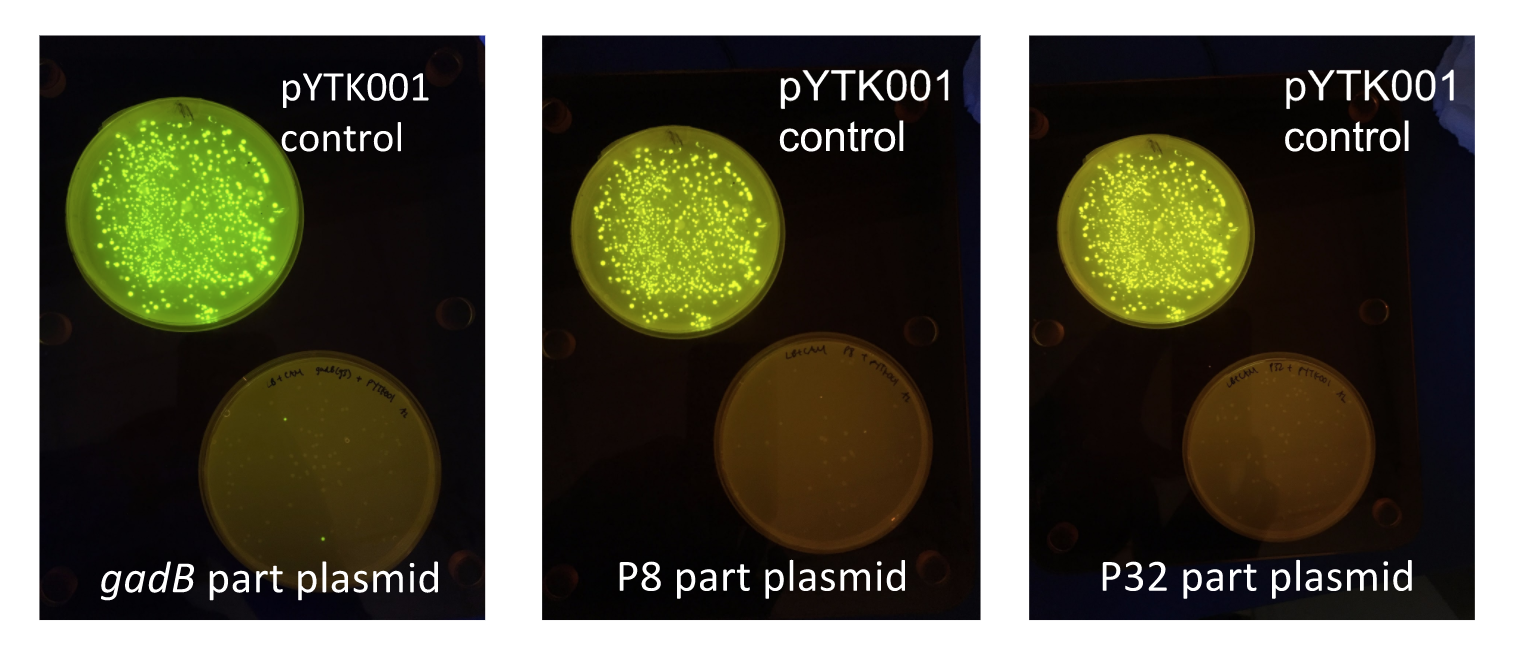
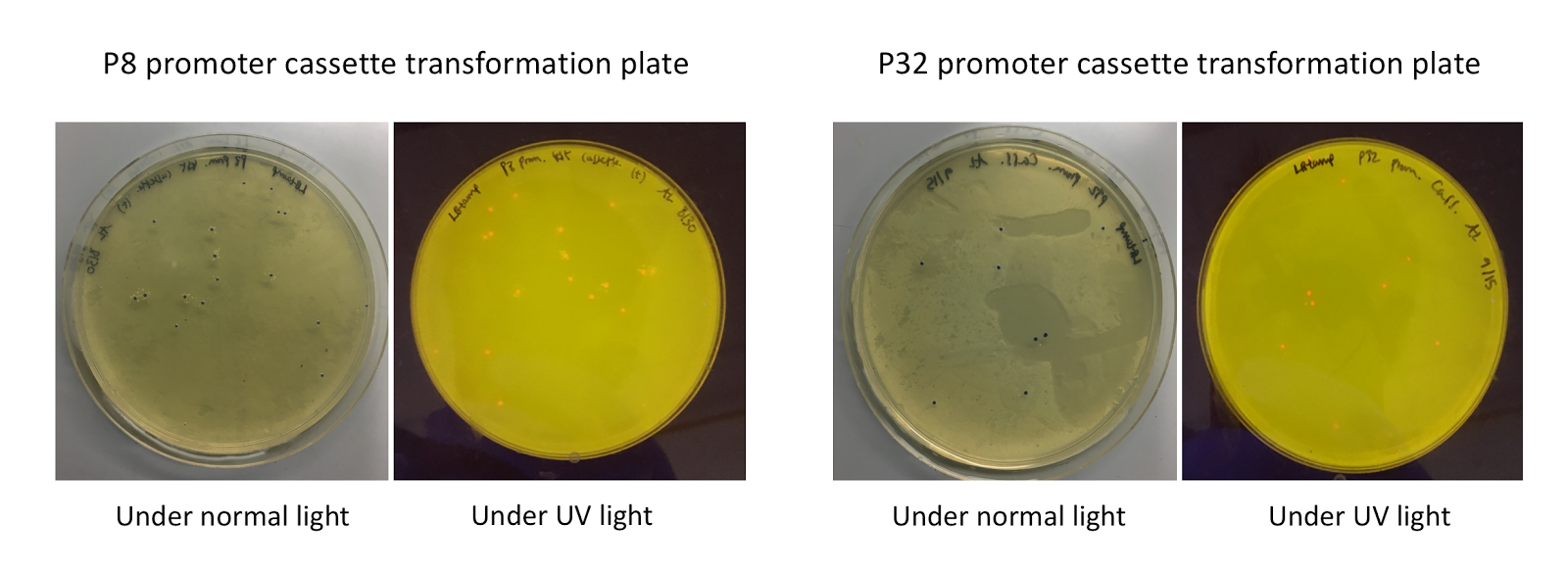
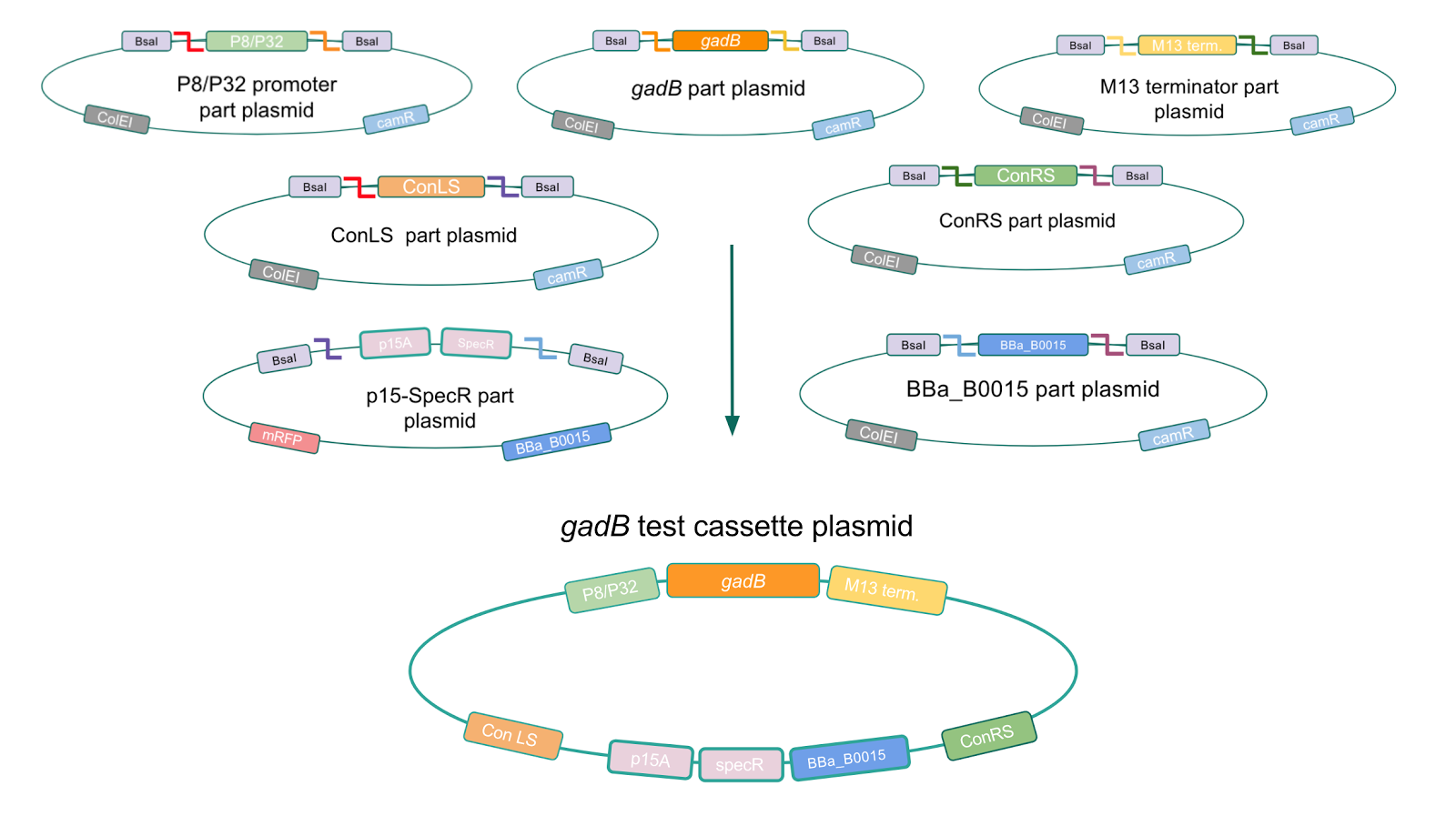
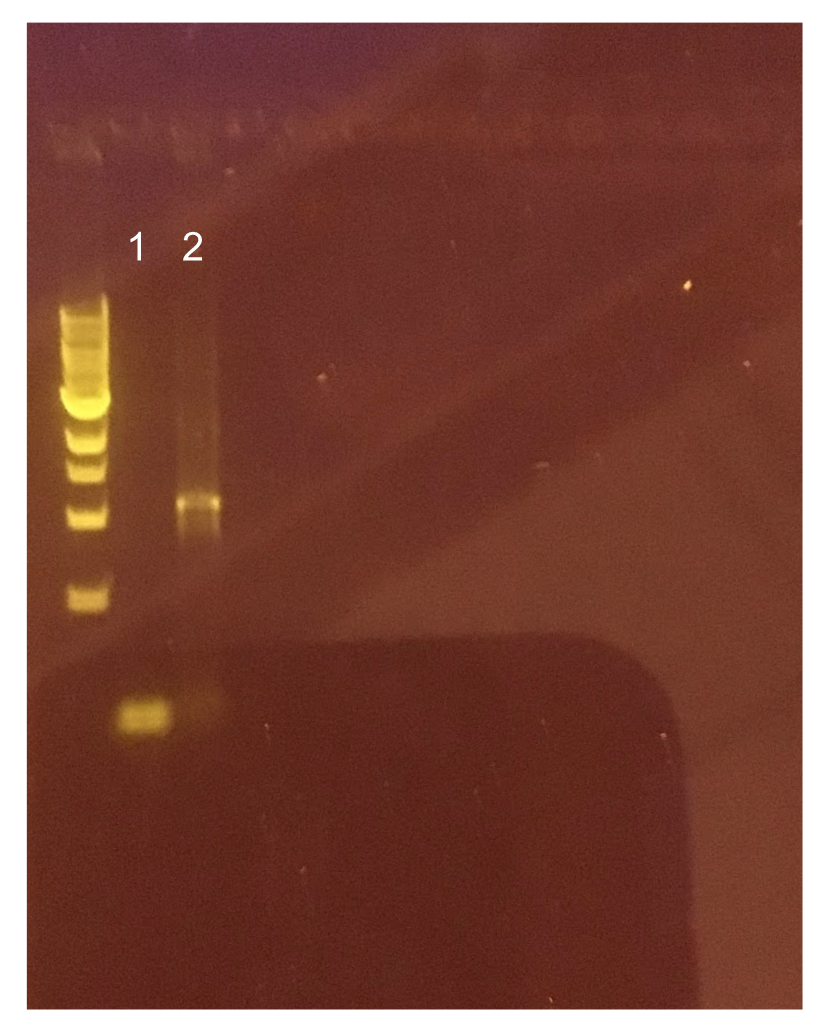
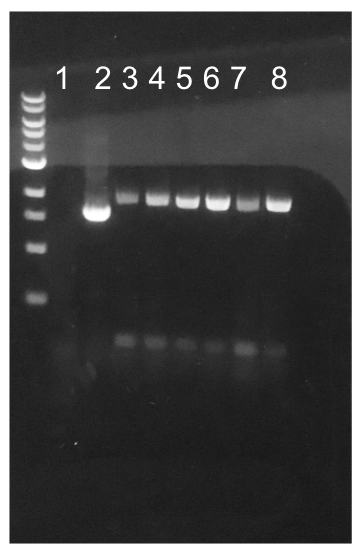
Similar to the pYTK001 entry vector in part assembly, the pYTK095 backbone used for cassette assembly contained a gfp reporter gene that is replaced by sequences of interest. This allowed us to easily perform a phenotypic screen for positive colonies. Non-fluorescent colonies may potentially have had the correct cassette assembly, while fluorescent colonies did not (Fig. 6). The non-fluorescent colonies were then screened using colony PCR (Fig. 7). The positive colonies were then miniprepped and sequenced. The sequencing results indicated that in all of the samples there were several point mutations within the gadB gene. We hypothesized that overexpression of the gadB gene via the P8 and P32 constitutive promoters and the high-copy number ColE1 origin induced a high metabolic load on the cells, resulting in mutational degradation of gadB. To troubleshoot this problem, we decided to use a backbone containing the low-copy number p15A origin for the cassette assembly (Fig. 8)
Creating a Golden Gate compatible vector
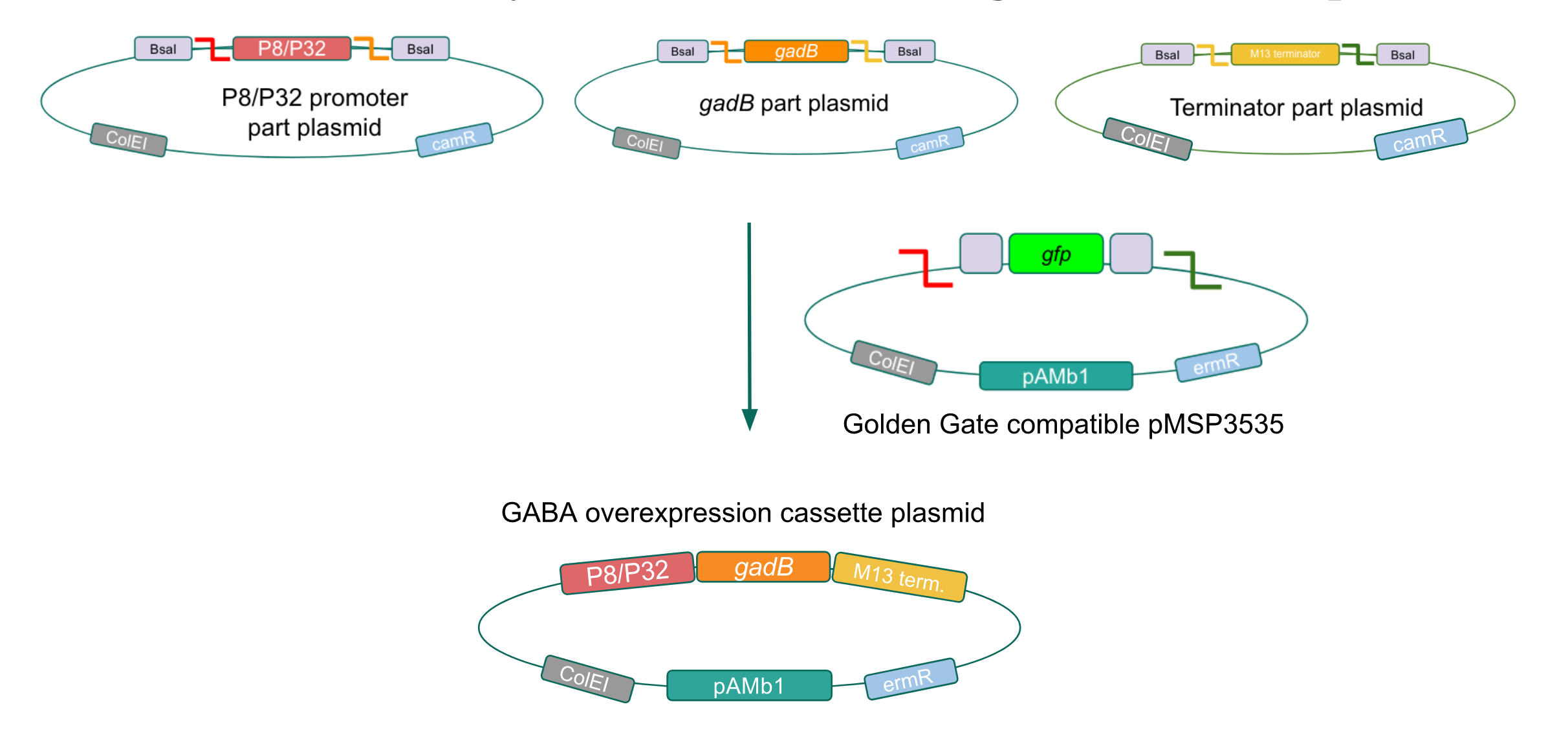
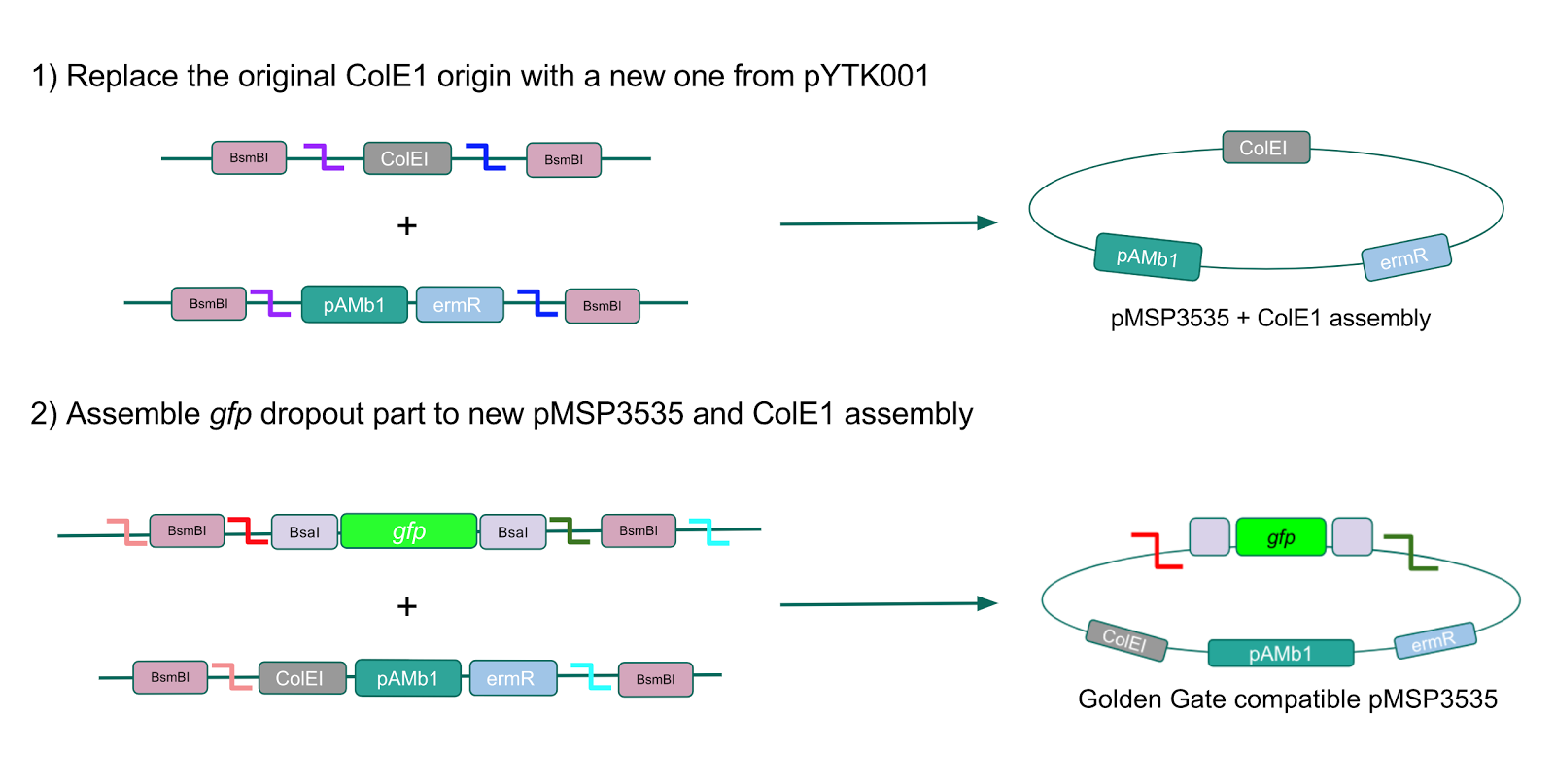
After confirming gadB overexpression in E. coli, we want to assemble our final GABA overexpression cassette plasmid using the vector pMSP3535 as the backbone (Fig. 9). To do this, we first needed to make pMSP3535 Golden Gate compatible (i.e. free of BsaI restriction sites and containing correct overhangs for cassette assembly). We chose to work with pMSP3535 as it contains both a ColE1 origin for replication in E. coli and a pAMb1 origin for replication in Gram-positive bacteria including Lactobacillus species (4). Additionally, the pMSP3535 vector contains the resistance gene for erythromycin, which Lactobacillus plantarum is naturally susceptible to (5)
The process of making the pMSP3535 vector Golden Gate compatible involved two steps: 1) assembling the pMSP3535 backbone (pAMb1 origin and erythromycin resistance gene) with a new ColE1 origin; 2) assembling a gfp dropout part to the assembly of the pMSP3535 backbone and the new ColE1 origin (Fig. 10).
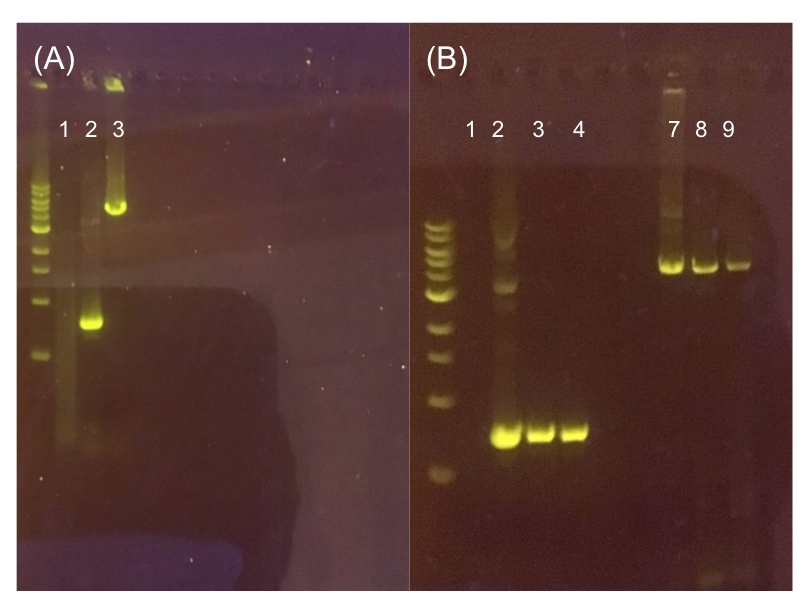
Since the original pMSP3535 vector contained two illegal BsaI sites within the ColE1 origin, we sought to replace this ColE1 origin with a BsaI-free one isolated from pYTK001. This assembly process involved linearizing and adding BsmBI sites and compatible overhangs to the pMSP3535 backbone and the pYTK001 ColE1 origin via PCR. After the pMSP3535 backbone and ColE1 origin were successfully amplified by PCR (Fig. 11a), they were joined together using BsmBI assembly. Diagnostic PCR was performed on pMSP3535 + ColE1 minipreps from E. coli transformants to screen for positive samples containing both the pMSP3535 backbone and the ColE1 inserts (Fig. 11b). After confirming the presence of the pMSP3535 vector and ColE1 origin, we partially sequence-confirmed the two miniprep samples.
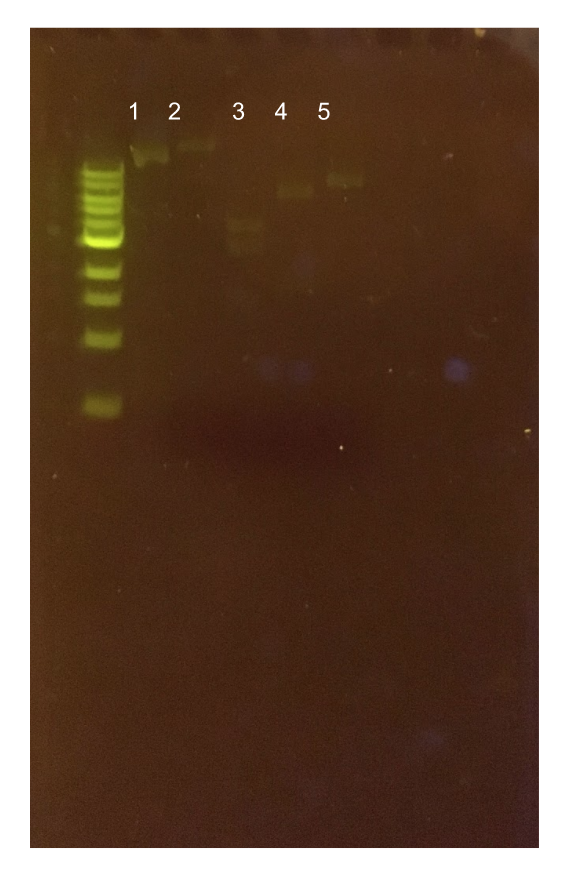
To this pMSP3535 + ColE1 assembly, we wanted to add a gfp dropout part containing internal BsaI sites that will generate overhangs compatible with those in the P8/P32 promoter and M13 terminator part plasmids. Additionally, the incorporation of this gfp dropout part will also allow us to visually screen for positive and negative transformants based on their fluorescence. BsmBI sites and compatible overhangs were added to the gfp dropout part by PCR amplifying it from pYTK047 (Fig. 12). We have been attempting to linearize and add BsmBI sites and overhangs to the positive pMSP3535 + ColE1 assemblies via PCR, with no success. However, results from diagnostic digests suggested that our assemblies may have contained extra, undesired DNA such as IS elements (Fig. 13). Thus, as of right now, we are screening for more positive pMSP3535 + ColE1 transformants. Once we have trouble-shooted this problem, the pMSP3535 + ColE1 and the gfp dropout PCR products will be joined through BsmBI assembly to form the final Golden Gate compatible pMSP3535 vector.
Assessing erythromycin susceptibility of E. coli
Since we are creating our Golden Gate compatible pMSP3535 shuttle vector in E. coli, we wanted to determine the natural susceptibility of E. coli to erythromycin as the minimum concentration to use has not been established clearly in the literature. Thus, we performed an erythromycin minimum inhibitory concentration test in liquid LB media (Fig. 14). After one-day incubation, we observed that E. coli was resistant up to around 150 µg/mL of erythromycin. From this experiment, we have determined that the optimal erythromycin concentration for selecting against E. coli in liquid culture is around 200-250 µg/mL.
References
- Yunes, R. A et al. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe. 42: 197-204 (2016).
- Zhu, D. et al. Isolation of strong constitutive promoters from Lactococcus lactis subsp. Lactis N8. FEMS Microbiol Lett. 363(16): pii: fnv107 (2015).
- Lee, M. E. et al. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 4(9): 975-86 (2015).
- Pérez-Arellano, I. et al. Construction of Compatible Wide-Host-Range Shuttle Vectors for Lactic Acid Bacteria and Escherichia coli. Plasmid. 46(2): 106-16 (2001).
- Flórez A. B. et al. Susceptibility of Lactobacillus plantarum Strains to Six Antibiotics and Definition of New Susceptibility–Resistance Cutoff Values. Microbial Drug Resistance. 12(4): 252-56 (2007).