| Line 296: | Line 296: | ||
<p style="font-family: verdana">Once we could successfully grow our chosen bacteria, we needed to transform the gram positive, <i> Lactobacillus plantarum</i>, with pMSP3535. In order to do this, we identified and worked with a different protocol than we had ever used in our lab. We attempted several protocols, including Landete 2014 (10) and Speer 2012 (11). However, we found success using a variation of the Welker protocol(12). Welker et al. transformed multiple strains of <i>Lactobacillus casei</i> using varying reagents and yielded different efficiencies between each strain of the species with each variation.</p> | <p style="font-family: verdana">Once we could successfully grow our chosen bacteria, we needed to transform the gram positive, <i> Lactobacillus plantarum</i>, with pMSP3535. In order to do this, we identified and worked with a different protocol than we had ever used in our lab. We attempted several protocols, including Landete 2014 (10) and Speer 2012 (11). However, we found success using a variation of the Welker protocol(12). Welker et al. transformed multiple strains of <i>Lactobacillus casei</i> using varying reagents and yielded different efficiencies between each strain of the species with each variation.</p> | ||
| − | + | <br> | |
<p style="font-family: verdana">The Texas Tech iGEM team helped us by testing the Speer protocol after we had attempted three procedures and hadn't successfully transformed. They were not able to successfully transform using the Speer protocol which suggests that the procedure was either too simplified for researchers who have never transformed gram-positive bacteria, or was not compatible with <i>Lactobacillus plantarum.</i> | <p style="font-family: verdana">The Texas Tech iGEM team helped us by testing the Speer protocol after we had attempted three procedures and hadn't successfully transformed. They were not able to successfully transform using the Speer protocol which suggests that the procedure was either too simplified for researchers who have never transformed gram-positive bacteria, or was not compatible with <i>Lactobacillus plantarum.</i> | ||
<button class="accordion">Electroporation of <i>L. plantarum</i></button> | <button class="accordion">Electroporation of <i>L. plantarum</i></button> | ||
| Line 314: | Line 314: | ||
<li>30% PEG 8000</li> | <li>30% PEG 8000</li> | ||
</ul> | </ul> | ||
| − | < | + | <br> |
<h4>Preparation of Recipient Cell Stocks</h4> | <h4>Preparation of Recipient Cell Stocks</h4> | ||
| + | <br> | ||
<ol style="font-family: verdana"> | <ol style="font-family: verdana"> | ||
<li>Inoculate 10-25 mL of -80 ࠷ L. Plantarum stocks in MRS broth at 37 ࠷ CO2 incubator, overnight without shaking.</li> | <li>Inoculate 10-25 mL of -80 ࠷ L. Plantarum stocks in MRS broth at 37 ࠷ CO2 incubator, overnight without shaking.</li> | ||
| Line 329: | Line 330: | ||
<li>Suspend cells in 0.5-0.6 mL of 30% PEG for storage at -80 ࠷. These should retain viability for up to 2 years.</li> | <li>Suspend cells in 0.5-0.6 mL of 30% PEG for storage at -80 ࠷. These should retain viability for up to 2 years.</li> | ||
</ol> | </ol> | ||
| + | <br> | ||
<h4>Treatment of Cells Prior to Electroporation</h4> | <h4>Treatment of Cells Prior to Electroporation</h4> | ||
<ol style="font-family: verdana"> | <ol style="font-family: verdana"> | ||
| Line 336: | Line 338: | ||
<li>After pretreatment, the cells were pelleted for 2-3 minutes in a microcentrifuge, washed once in 1 mL of cold, sterile, 30% PEG solution and suspended in 0.5-0.6 mL cold, sterile PEG solution for electroporation.</li> | <li>After pretreatment, the cells were pelleted for 2-3 minutes in a microcentrifuge, washed once in 1 mL of cold, sterile, 30% PEG solution and suspended in 0.5-0.6 mL cold, sterile PEG solution for electroporation.</li> | ||
</ol> | </ol> | ||
| + | <br> | ||
<h4 style="font-family: verdana">Electroporation</h4> | <h4 style="font-family: verdana">Electroporation</h4> | ||
<ol style="font-family: verdana"> | <ol style="font-family: verdana"> | ||
| Line 346: | Line 349: | ||
<li>Cells stored in glycerol (-80 ࠷) after electroporation incubation can be expected to yield similar results to the cells that were plated directly after the incubation.</li> | <li>Cells stored in glycerol (-80 ࠷) after electroporation incubation can be expected to yield similar results to the cells that were plated directly after the incubation.</li> | ||
</ol> | </ol> | ||
| − | + | <br> | |
</div> | </div> | ||
| Line 352: | Line 355: | ||
<p style="font-family: verdana">After preparing the necessary solutions, we followed the Welker protocol with some minor differences. We inoculated bacterial stocks with 10 mL of MRS broth in a CO2 incubator, without shaking, overnight. After this, we subcultured the bacteria in 200 mL of prewarmed MRS broth with 0.9M NaCl from an OD600 of 0.1 until it reached an OD600 of 0.6 (~6 hours). After harvesting the cells by centrifugation and rinsing, we resuspended the cells in 4 mL of cold water and eight 0.5 mL aliquots were divided into 1.5 mL microcentrifuge tubes.</p> | <p style="font-family: verdana">After preparing the necessary solutions, we followed the Welker protocol with some minor differences. We inoculated bacterial stocks with 10 mL of MRS broth in a CO2 incubator, without shaking, overnight. After this, we subcultured the bacteria in 200 mL of prewarmed MRS broth with 0.9M NaCl from an OD600 of 0.1 until it reached an OD600 of 0.6 (~6 hours). After harvesting the cells by centrifugation and rinsing, we resuspended the cells in 4 mL of cold water and eight 0.5 mL aliquots were divided into 1.5 mL microcentrifuge tubes.</p> | ||
| − | + | <br> | |
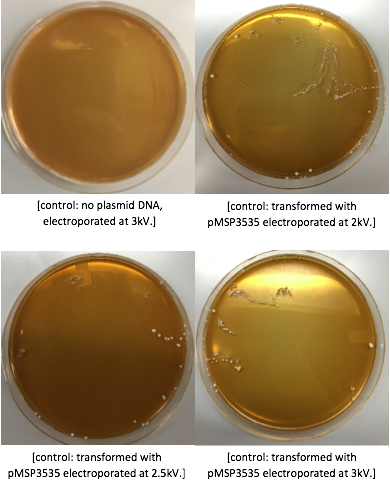
<p style="font-family: verdana">The cells were washed several more times with water and 30% PEG 8000, then stored in a -80°C freezer. These cells should stay viable for up to two years.To enhance the transformation efficiency, 600 μL of prepared cells were suspended 900 μL of cold, sterile, distilled water for 30 min as a pretreatment to electroporation.The cells were pelleted and washed several times with water and 30% PEG 8000. We added 100 ng of plasmid DNA to 100μL of cell suspension as opposed to 200 ng that the protocol recommends; our first plasmid of choice was pMSP3535. This plasmid expresses erythromycin resistance. The cells were electroporated in 2mm cuvettes using the Ec3 settings on the BioRad electroporator which corresponds to 600Ω and 3 kV, differing from the Welker protocol recommendations. | <p style="font-family: verdana">The cells were washed several more times with water and 30% PEG 8000, then stored in a -80°C freezer. These cells should stay viable for up to two years.To enhance the transformation efficiency, 600 μL of prepared cells were suspended 900 μL of cold, sterile, distilled water for 30 min as a pretreatment to electroporation.The cells were pelleted and washed several times with water and 30% PEG 8000. We added 100 ng of plasmid DNA to 100μL of cell suspension as opposed to 200 ng that the protocol recommends; our first plasmid of choice was pMSP3535. This plasmid expresses erythromycin resistance. The cells were electroporated in 2mm cuvettes using the Ec3 settings on the BioRad electroporator which corresponds to 600Ω and 3 kV, differing from the Welker protocol recommendations. | ||
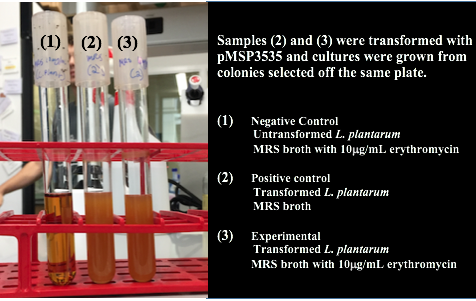
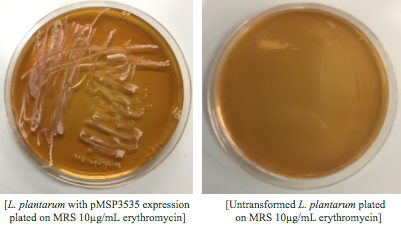
After cells were electroporated, they recovered overnight in the appropriate recovery media. They were plated on MRS agar plates with 10μg/mL erythromycin and left to grow for 2 days. </p> | After cells were electroporated, they recovered overnight in the appropriate recovery media. They were plated on MRS agar plates with 10μg/mL erythromycin and left to grow for 2 days. </p> | ||
| − | + | <br> | |
</html> | </html> | ||
[[File:T--Austin_UTexas--LPlantarumTrans.jpg|thumb|center|800px]] | [[File:T--Austin_UTexas--LPlantarumTrans.jpg|thumb|center|800px]] | ||
<html> | <html> | ||
| − | + | <br> | |
<p style="font-family: verdana">Colonies from each transformation plate were grown up in MRS broth supplemented with 10μg/mL erythromycin. The subsequent day, they were streaked on 10μg/mL erythromycin MRS agar plates to verify resistance.</p> | <p style="font-family: verdana">Colonies from each transformation plate were grown up in MRS broth supplemented with 10μg/mL erythromycin. The subsequent day, they were streaked on 10μg/mL erythromycin MRS agar plates to verify resistance.</p> | ||
| − | + | <br> | |
</html> | </html> | ||
[[File:T--Austin_UTexas--LPlantarumExp.jpg|thumb|center|800px]] | [[File:T--Austin_UTexas--LPlantarumExp.jpg|thumb|center|800px]] | ||
<html> | <html> | ||
| − | + | <br> | |
</html> | </html> | ||
[[File:T--Austin_UTexas--LPlantarumPlates.jpg|thumb|center|800px]] | [[File:T--Austin_UTexas--LPlantarumPlates.jpg|thumb|center|800px]] | ||
<html> | <html> | ||
| − | + | <br> | |
<p style="font-family: verdana">We aimed to show a visible transformation of <i>Lactobacillus plantarum</i> in addition to the resistance, so we transformed again using a different plasmid. This time we transformed with pBTK501, a plasmid that codes for a resistance to ampicillin and expresses <i>gfp</i>.</p> | <p style="font-family: verdana">We aimed to show a visible transformation of <i>Lactobacillus plantarum</i> in addition to the resistance, so we transformed again using a different plasmid. This time we transformed with pBTK501, a plasmid that codes for a resistance to ampicillin and expresses <i>gfp</i>.</p> | ||
| − | + | <br> | |
<h2 style="font-family: verdana; font-size: 30px; text-align: center">References</h2> | <h2 style="font-family: verdana; font-size: 30px; text-align: center">References</h2> | ||
<ol style="font-size:13px; font-family: verdana"> | <ol style="font-size:13px; font-family: verdana"> | ||
Revision as of 06:51, 1 November 2017
Click on one of the images below to learn more about our results!
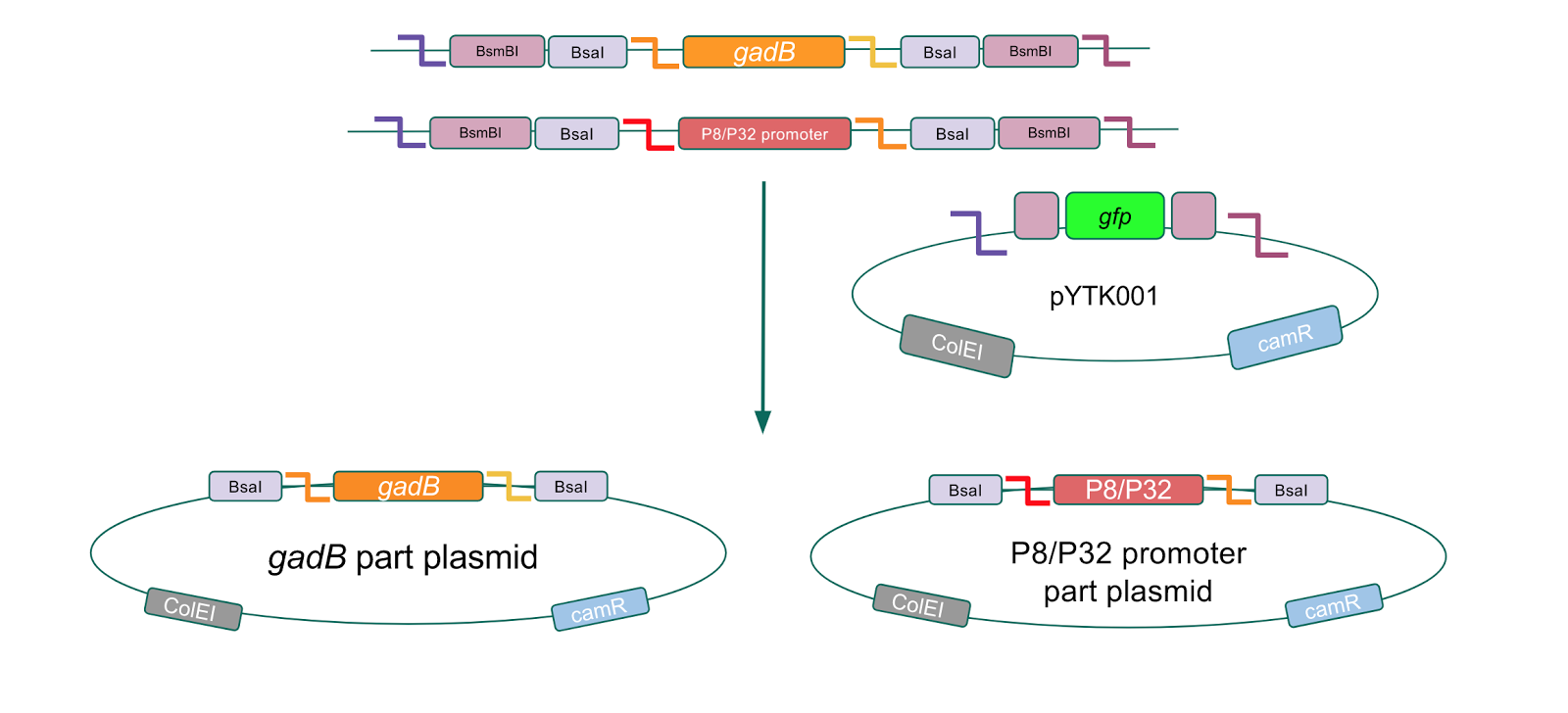
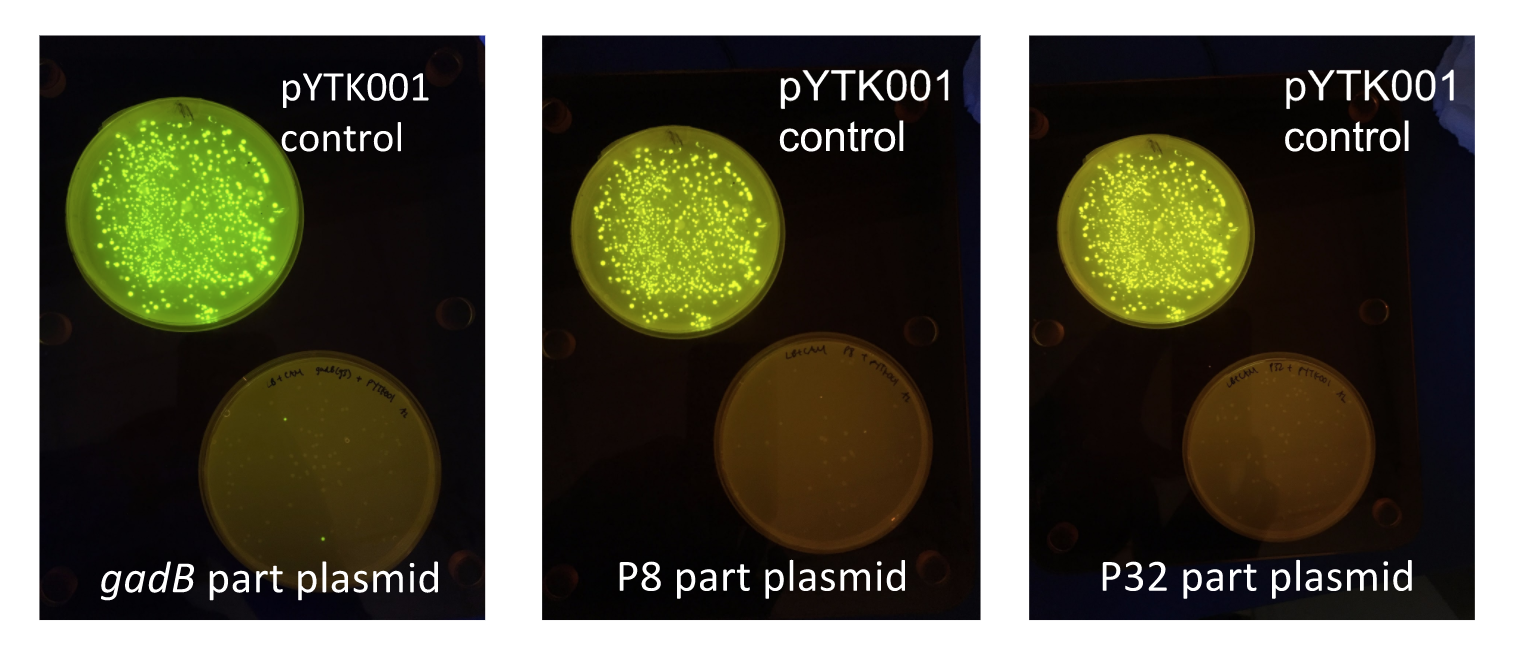
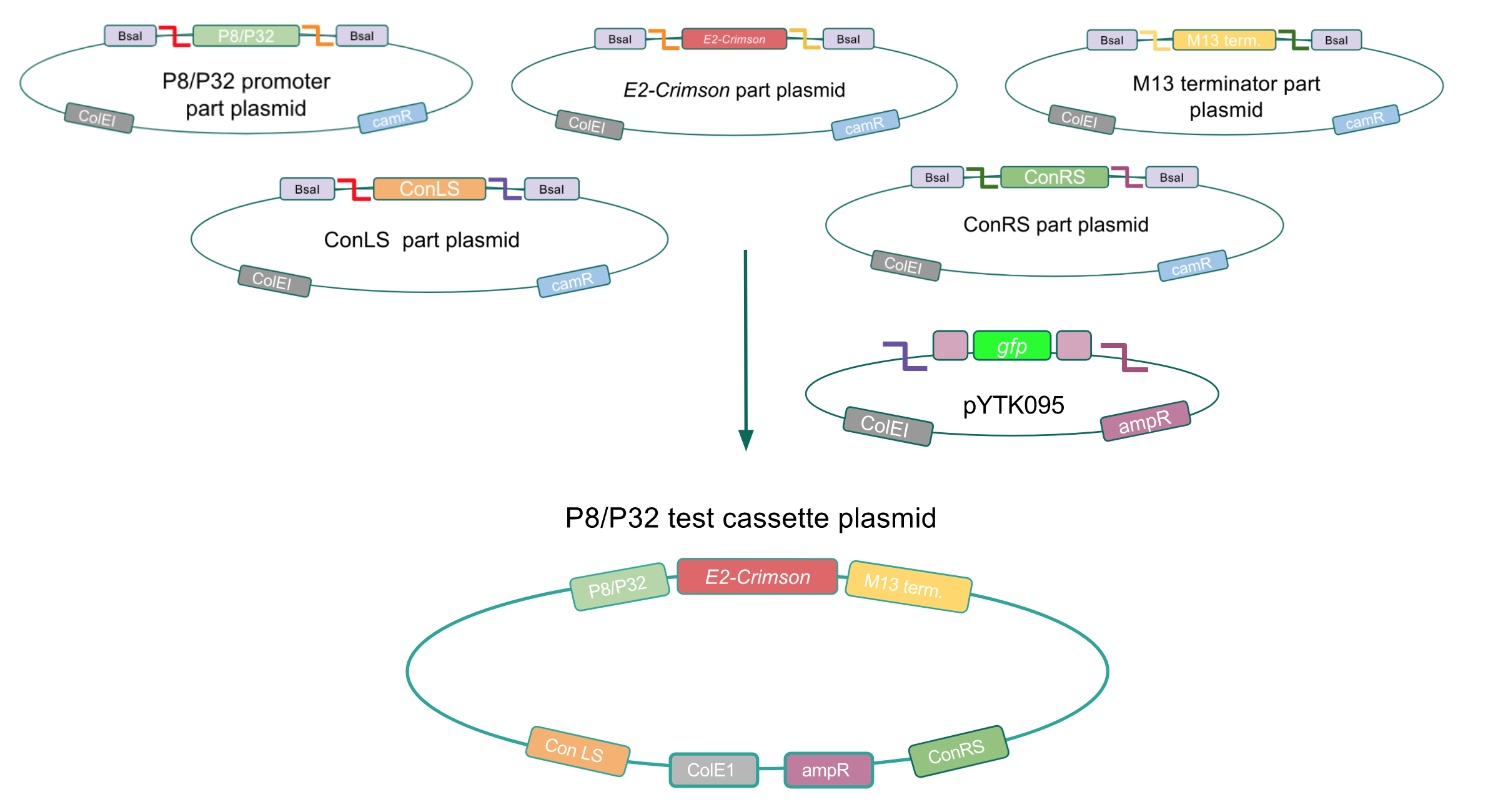
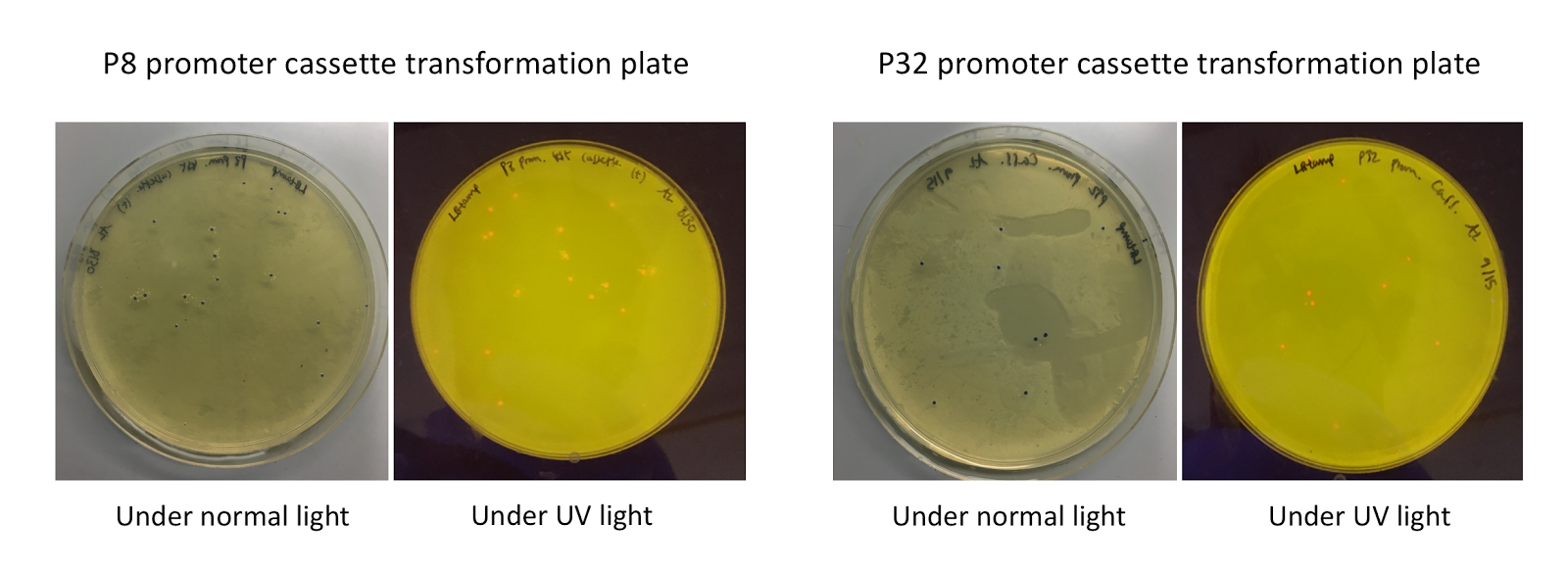
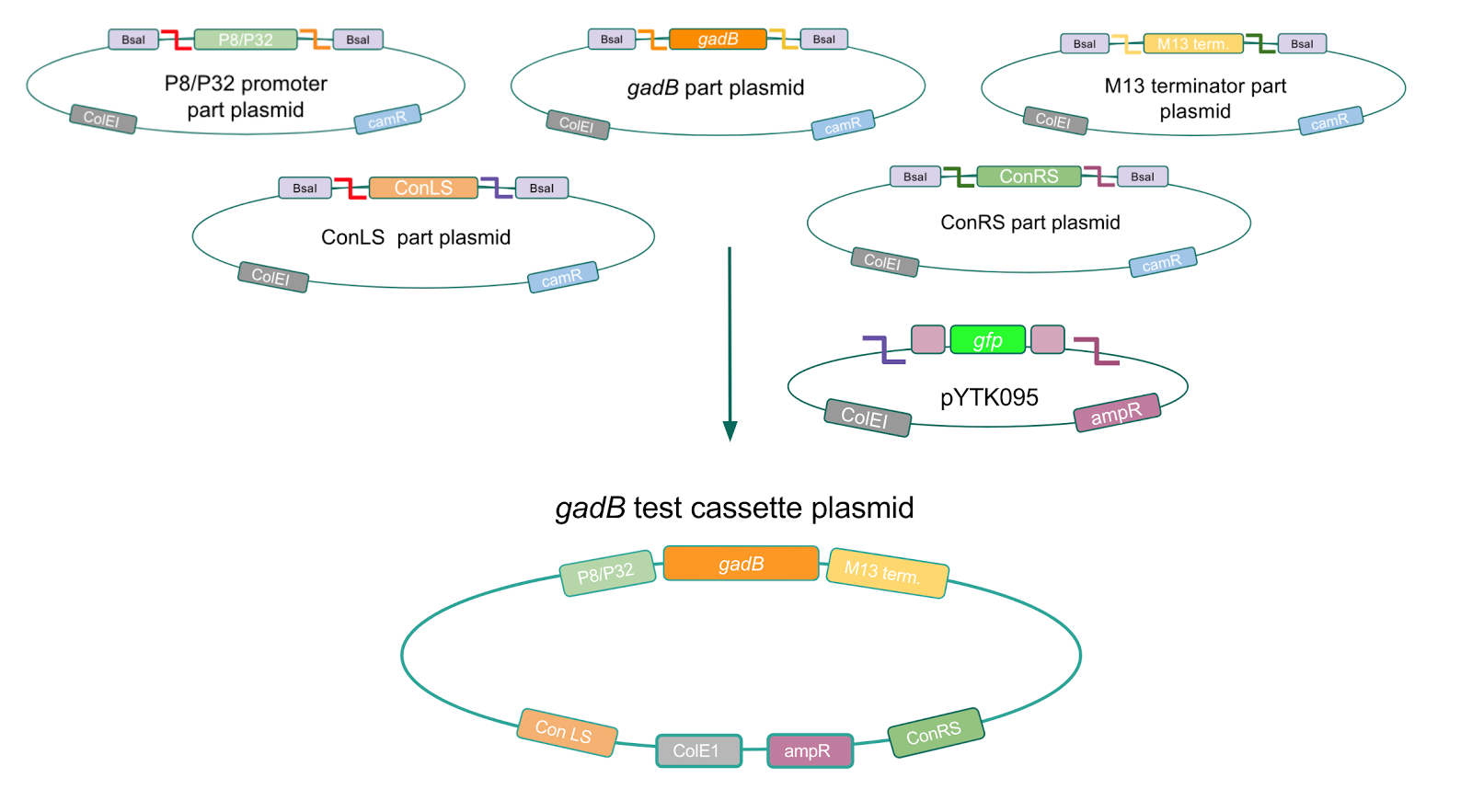
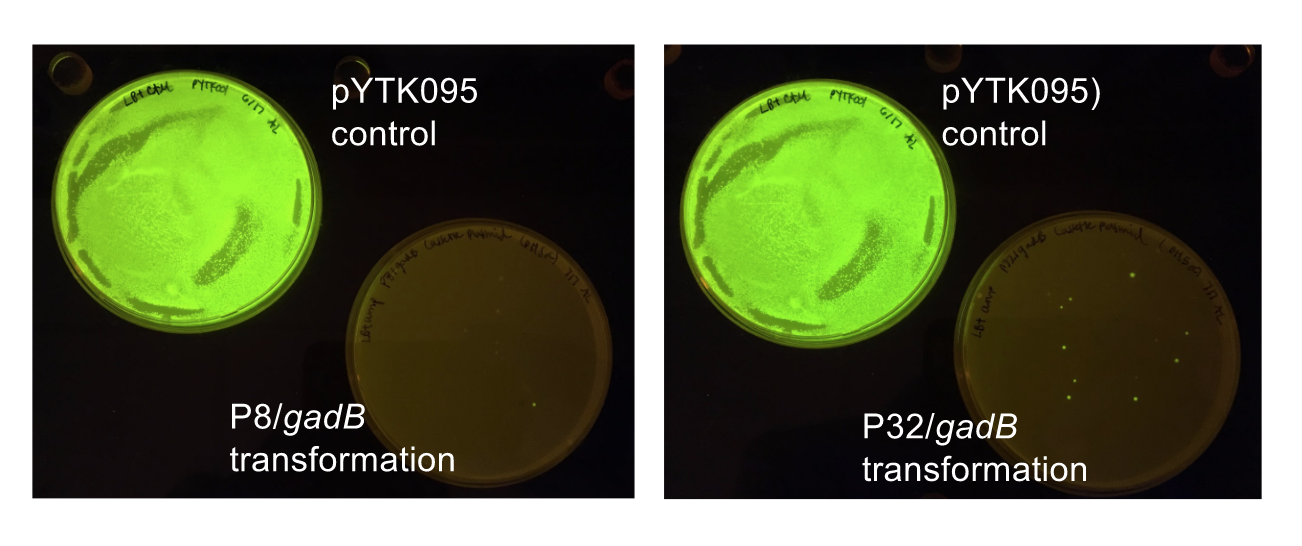
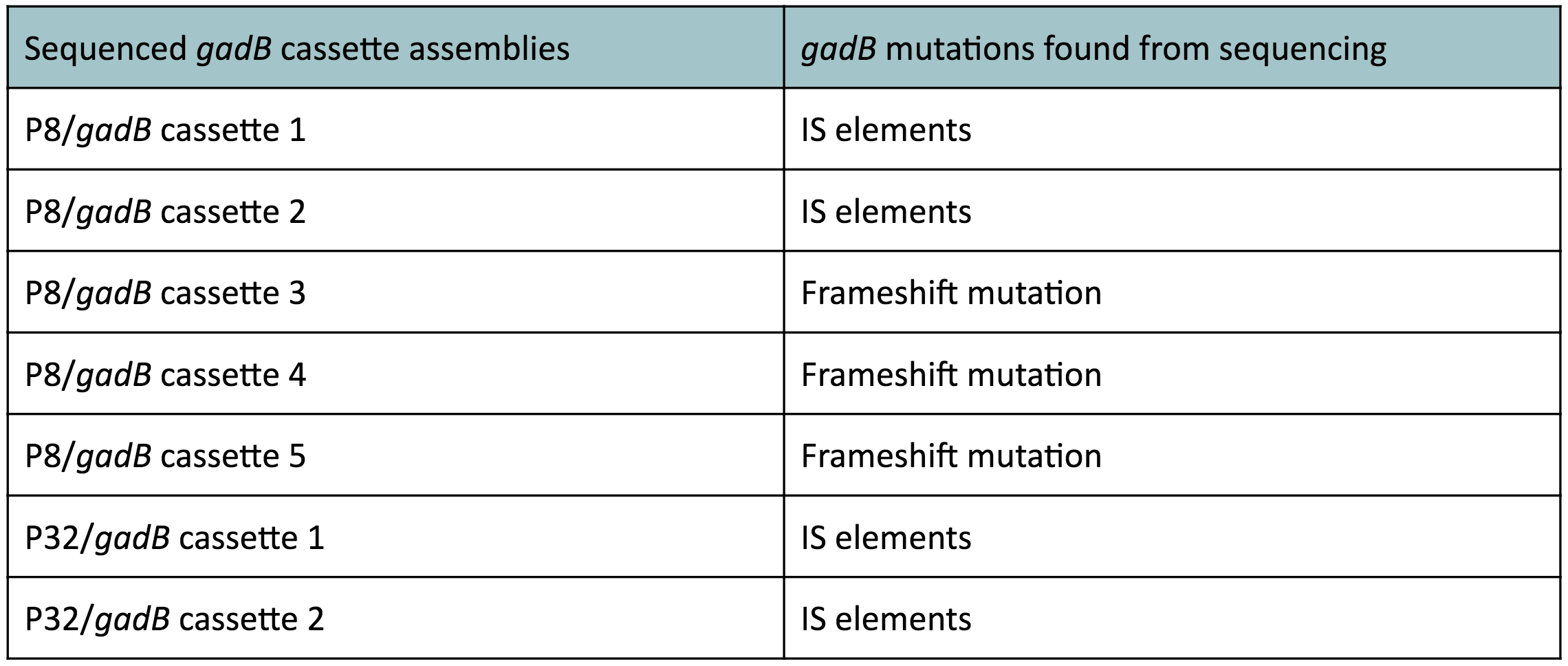
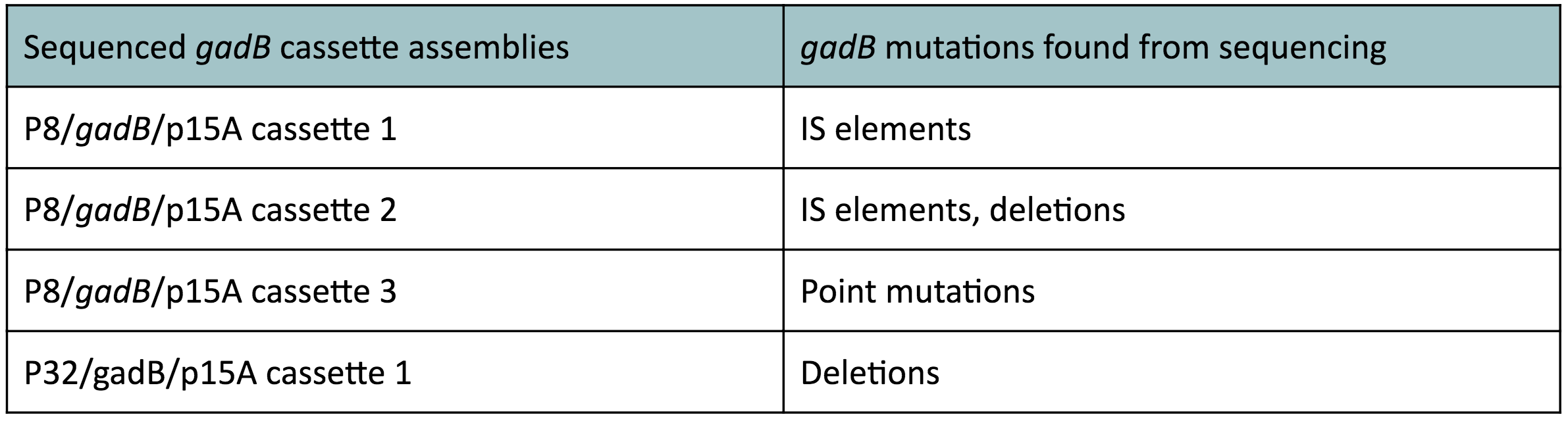
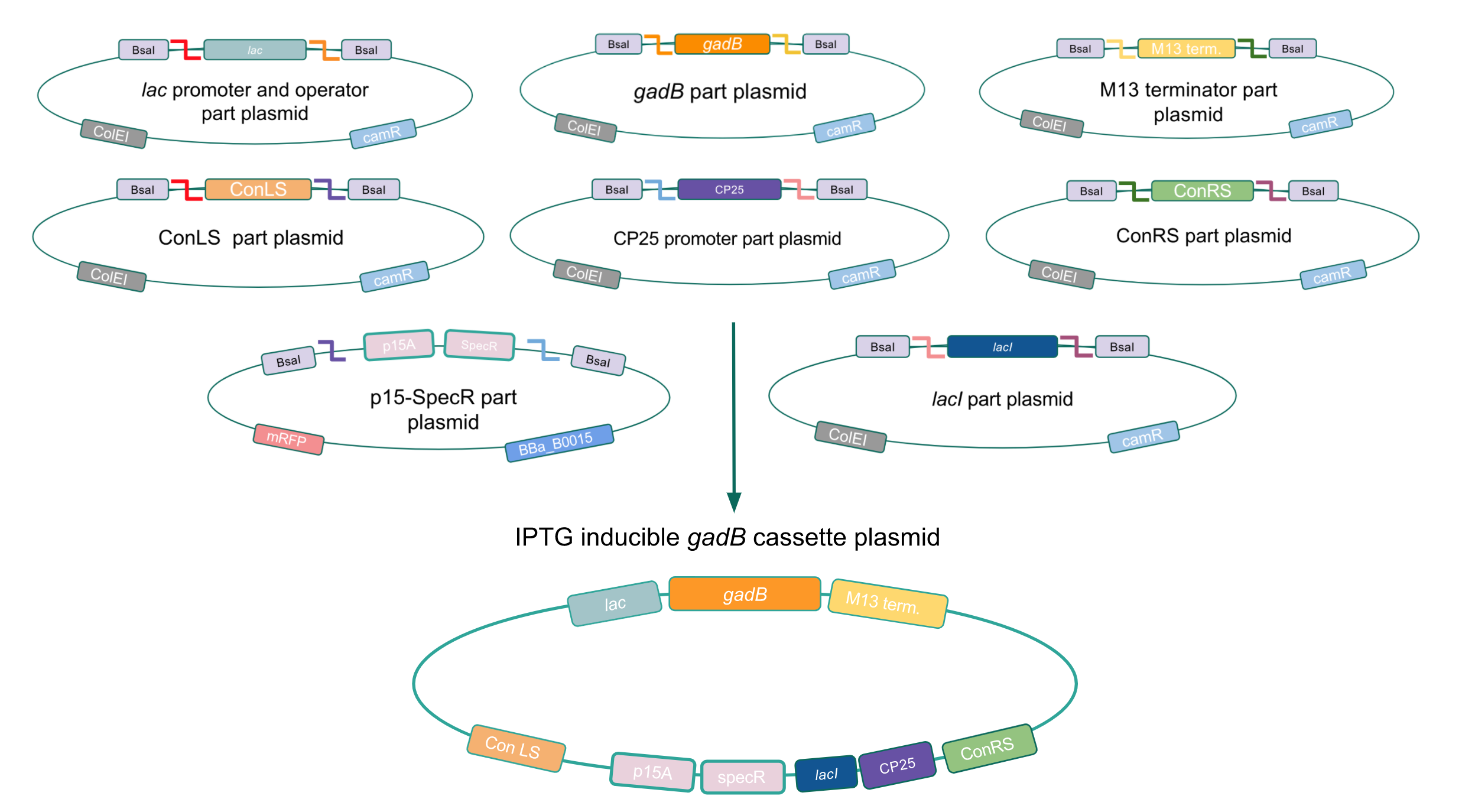
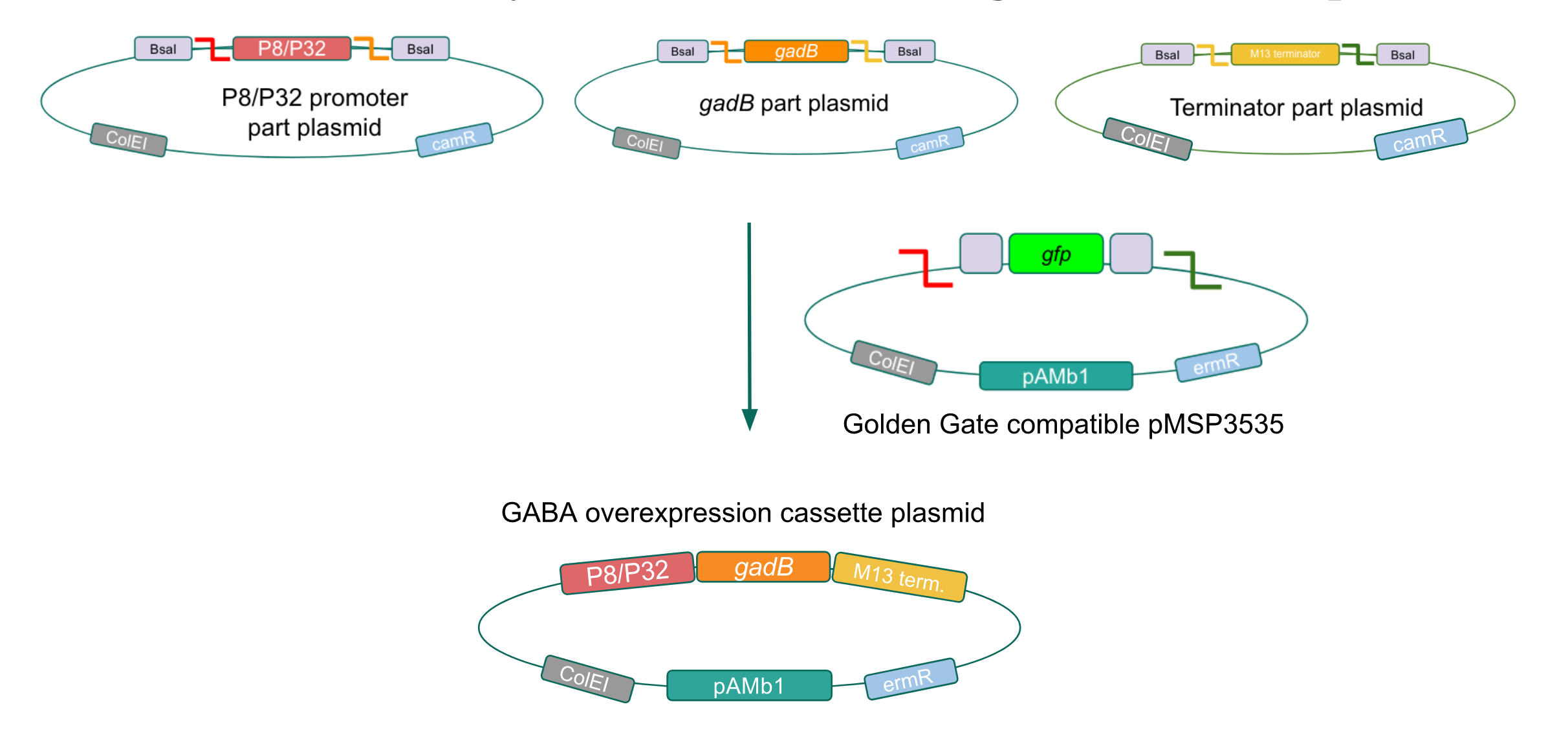
Although bacteria can naturally synthesize GABA, we wanted to increase expression of the gadB gene and subsequently GABA production in order to imbue our probiotic with a more potent medicinal quality, with the idea that this GABA-overproducing probiotic can then be consumed by patients with bowel disorders or anxiety (1). Overexpression of the gadB gene will be accomplished by placing it under the control of either the P8 or P32 constitutive promoters from Lactococcus lactis (2).
To make our GABA-producing probiotic we first needed to assemble a GABA overexpression cassette plasmid using the Golden Gate assembly method. The intention here is that bacteria containing this GABA overexpression cassette plasmid should produce high levels of GABA. In short, Golden Gate Assembly is a new cloning method that allows for the creation of a multi-part DNA assembly (i.e. cassette plasmid) in a single reaction through the use of DNA parts containing specific, predefined suffixes and prefixes with recognition sites for Type IIs restriction enzymes (e.g. BsmBI and BsaI). The specificity of these suffixes and prefixes provides directionality of the desired DNA parts during the assembly process. For our purposes, we used the MoClo Yeast Tool Kit developed by John Dueber (3).
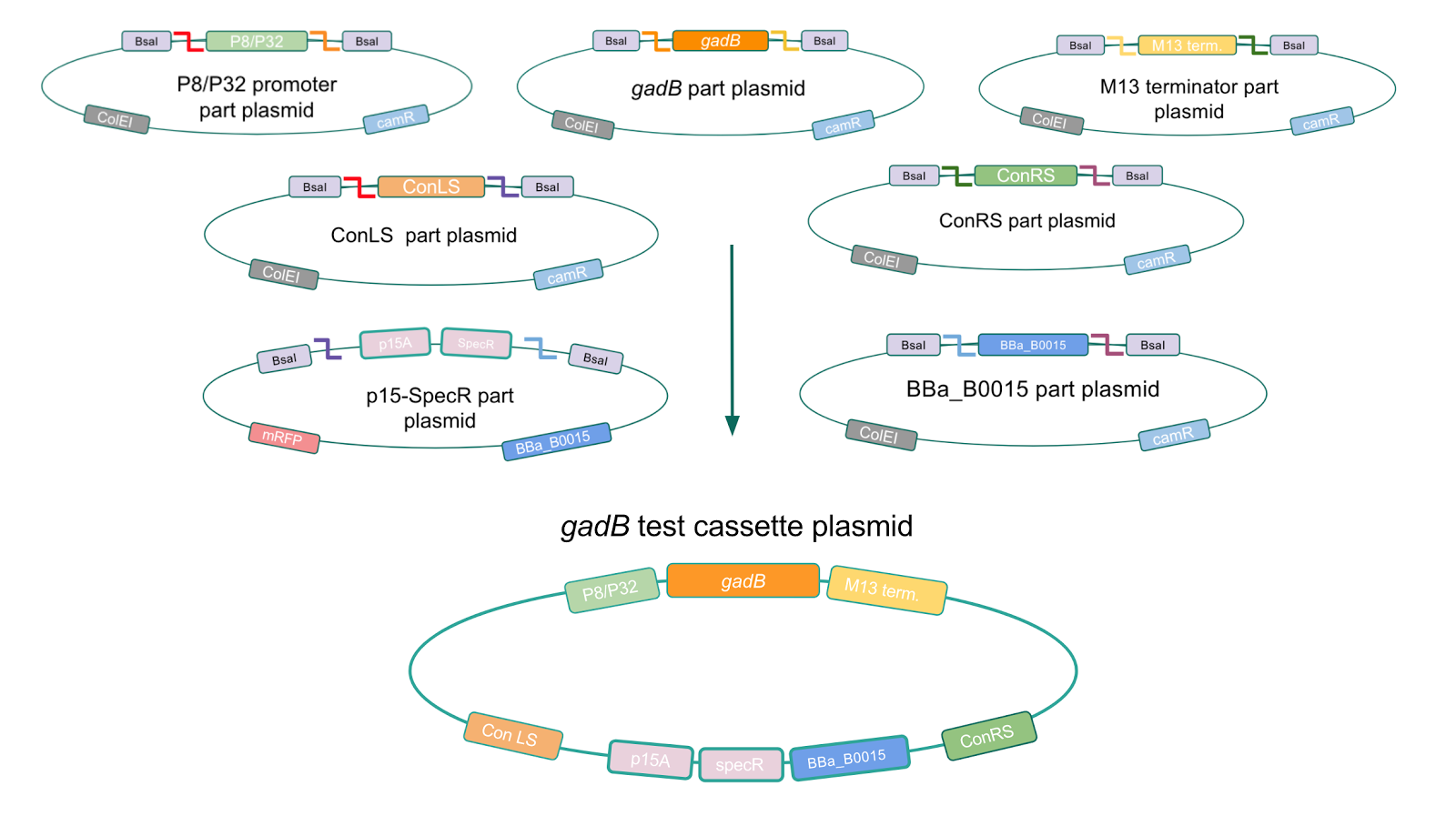
Creating a Golden Gate compatible shuttle vector
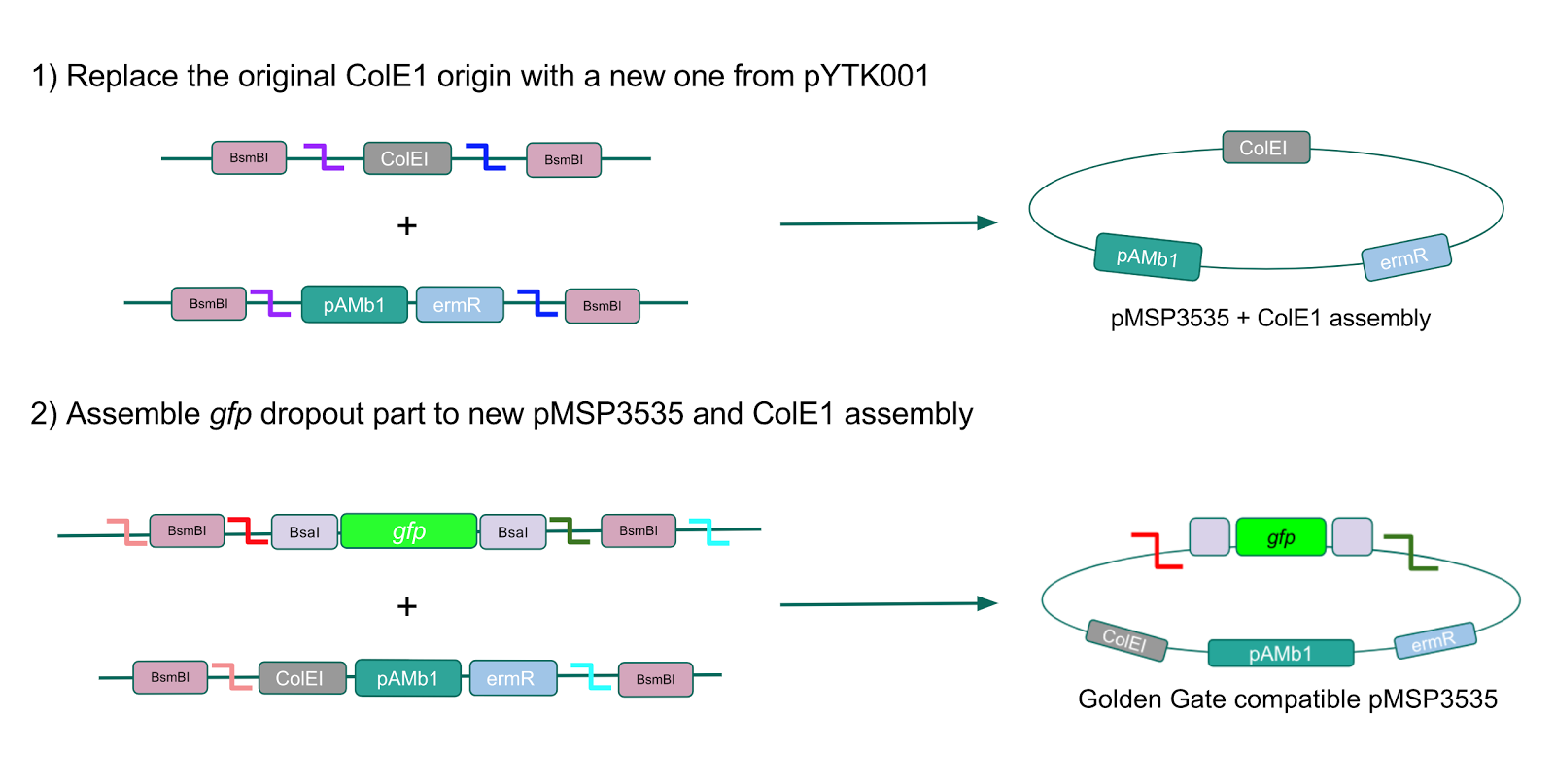
We wanted to assemble our final GABA overexpression cassette plasmid using the shuttle vector pMSP3535 as the backbone (Fig. 9). To do this, we first needed to make pMSP3535 Golden Gate compatible (i.e. free of BsaI restriction sites and containing correct overhangs for cassette assembly). We chose to work with pMSP3535 as it contains both a ColE1 origin for replication in E. coli and a pAMb1 origin for replication in Gram-positive bacteria including Lactobacillus species (6). Additionally, the pMSP3535 vector contains the resistance gene for erythromycin, of which Lactobacillus plantarum is naturally susceptible (7).
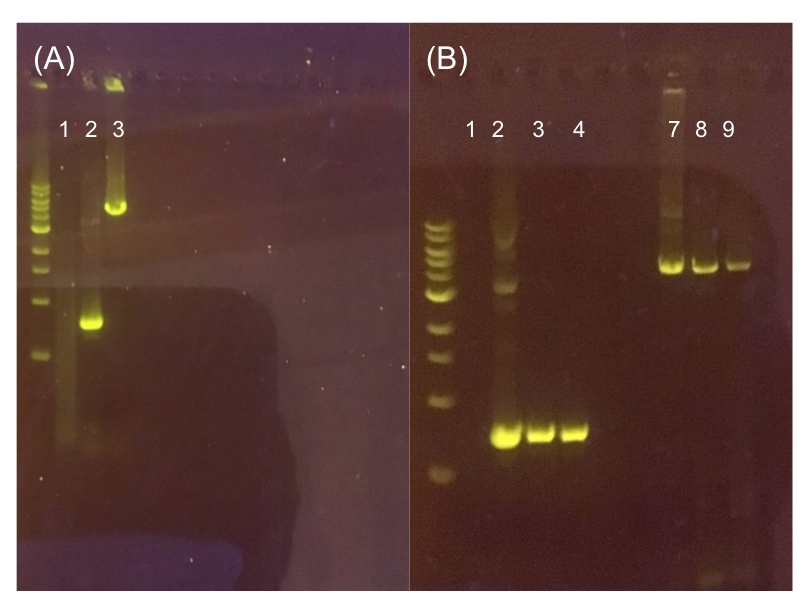
The process of making the pMSP3535 vector Golden Gate compatible involved two steps: 1) assembling the pMSP3535 backbone (pAMb1 origin and erythromycin resistance gene) with a new ColE1 origin; 2) assembling a gfp dropout part to the assembly of the pMSP3535 backbone and the new ColE1 origin (Fig. 10).
Assessing erythromycin susceptibility of E. coli
Because we are creating our Golden Gate compatible pMSP3535 shuttle vector in E. coli, we wanted to determine the natural susceptibility of E. coli to erythromycin as the minimum concentration to use has not been established clearly in the literature. Thus, we performed an erythromycin minimum inhibitory concentration test in liquid LB media (Fig. 13). After one-day incubation, we observed that E. coli was resistant up to around 150 µg/mL of erythromycin. From this experiment, we have determined that the optimal erythromycin concentration for selecting against E. coli in liquid culture is around 200-250 µg/mL.