| Line 9: | Line 9: | ||
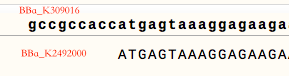
To indicate neuron AWB in C.elegans’ head, we chose GFP for <i>Caenorhabditis elegans</i>. It’s the part BBa_K309016 submitted by iGEM10_Queens-Canada. However, the sample status shows that ”it’s complicated”, which means the sample may not have been sequenced or there is some problems on it. | To indicate neuron AWB in C.elegans’ head, we chose GFP for <i>Caenorhabditis elegans</i>. It’s the part BBa_K309016 submitted by iGEM10_Queens-Canada. However, the sample status shows that ”it’s complicated”, which means the sample may not have been sequenced or there is some problems on it. | ||
| − | {{SUSTech_Image_fill-width | filename=T--SUSTech_Shenzhen--Charicterization1.png|width= | + | {{SUSTech_Image_fill-width | filename=T--SUSTech_Shenzhen--Charicterization1.png|width=1000px|caption=<B>Head of <i>C.elegans</i></B>}} |
| + | |||
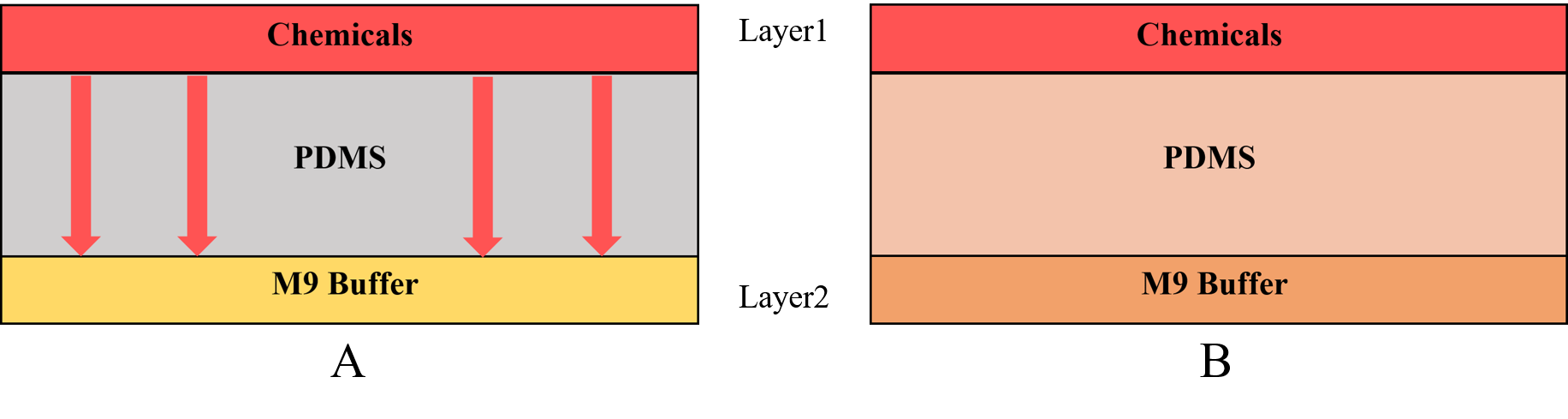
| + | {{SUSTech_Image_Center_fill-width | filename=T--SUSTech_Shenzhen--Microfuildics--layer.png|width=1000px|caption=<B>Fig.3 A) The chemicals diffuse downward across the PDMS. B) The chemical can across the PDMS to enter in the M9 buffer (the buffer for worms).</B> The M9 buffer is injected into the layer2 continually so the chemicals will not be accumulated in the M9 buffer, on the other words the concentration of layer2 is always equal to the concentration at the bottom of the PDMS.}} | ||
| + | |||
To use GFP for C.elegans, we needed to modify and confirm this GFP. After getting sequence of BBa_K309016 sequence, we find that the part doesn’t start with start codon “ATG". We optimize it by deleting 9bp in front of the start codon make the coding sequence starts with “ATG”. | To use GFP for C.elegans, we needed to modify and confirm this GFP. After getting sequence of BBa_K309016 sequence, we find that the part doesn’t start with start codon “ATG". We optimize it by deleting 9bp in front of the start codon make the coding sequence starts with “ATG”. | ||
Revision as of 09:36, 27 October 2017
Characterization
Parts
To indicate neuron AWB in C.elegans’ head, we chose GFP for Caenorhabditis elegans. It’s the part BBa_K309016 submitted by iGEM10_Queens-Canada. However, the sample status shows that ”it’s complicated”, which means the sample may not have been sequenced or there is some problems on it.
Template:SUSTech Image fill-width
To use GFP for C.elegans, we needed to modify and confirm this GFP. After getting sequence of BBa_K309016 sequence, we find that the part doesn’t start with start codon “ATG". We optimize it by deleting 9bp in front of the start codon make the coding sequence starts with “ATG”.
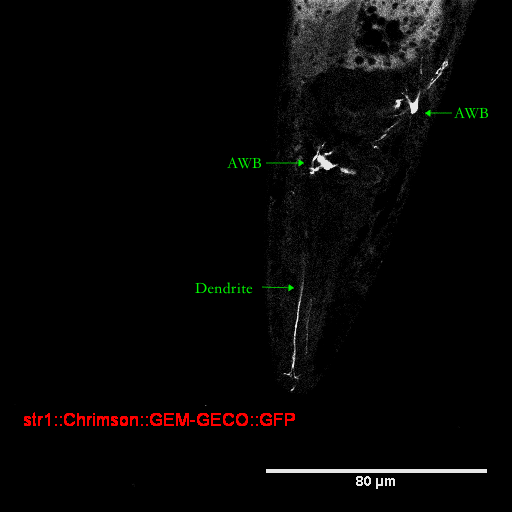
We successfully improved characterization of GFP for C.elegans. We submit this optimized GFP as our part BBa_K2492000. We constructed a str-1::Chrimson::GEMgeco::GFP fusion gene and injected it into C.elegans. Promoter str-1 drives the expression of GFP in neuron AWB, which indicate the AWB location.
We capture the image of GFP specifically expressed in AWB with confocal microscope and clearly observe AWB with its dendrite(Figure).