| Line 285: | Line 285: | ||
<p style="font-family: verdana"><i> Lactobacillus plantarum </i> is a gram-positive lactic acid producing bacteria, so it requires a different growth media than we typically use in our lab. In 1954, Briggs agar was developed (8). This media was designed for lactobacilli, but was not sufficient for many species, including <i> Lactobacillus plantarum </i>, so a different non-selective media for general lactobacilli was developed in 1960 by Man, Rogosa and Sharpe and named MRS (9). We have exclusively grown our <i> Lactobacillus plantarum </i> on MRS media. Further, we grew <i> Lactobacillus plantarum </i> in a CO2 incubator as referenced in most literature we studied (10-12). The metabolic pathways in the bacteria alters when grown aerobically to produce excess acetate (13) and less lactic acid. Because we intend to utilize this bacteria in a fermentable food, a change in this metabolic pathway would not benefit our ultimate goal.</p> | <p style="font-family: verdana"><i> Lactobacillus plantarum </i> is a gram-positive lactic acid producing bacteria, so it requires a different growth media than we typically use in our lab. In 1954, Briggs agar was developed (8). This media was designed for lactobacilli, but was not sufficient for many species, including <i> Lactobacillus plantarum </i>, so a different non-selective media for general lactobacilli was developed in 1960 by Man, Rogosa and Sharpe and named MRS (9). We have exclusively grown our <i> Lactobacillus plantarum </i> on MRS media. Further, we grew <i> Lactobacillus plantarum </i> in a CO2 incubator as referenced in most literature we studied (10-12). The metabolic pathways in the bacteria alters when grown aerobically to produce excess acetate (13) and less lactic acid. Because we intend to utilize this bacteria in a fermentable food, a change in this metabolic pathway would not benefit our ultimate goal.</p> | ||
| − | + | <br> | |
<button class="accordion">MRS Media</button> | <button class="accordion">MRS Media</button> | ||
<div class="panel"> | <div class="panel"> | ||
| + | <br> | ||
<ul style="font-family: verdana"> | <ul style="font-family: verdana"> | ||
<li>MRS broth was prepared using 55g of Difco MRS broth powder in 1L of distilled water. The solution was then autoclaved and stored in a 4 degree refrigerator along with the Difco MRS powder.</li> | <li>MRS broth was prepared using 55g of Difco MRS broth powder in 1L of distilled water. The solution was then autoclaved and stored in a 4 degree refrigerator along with the Difco MRS powder.</li> | ||
<li>MRS agar was prepared by mixing 55g of Difco MRS broth powder and 15g of agar flakes in 1 L of water. The solution was then autoclaved and stored in a 4 degree refrigerator along with the Difco MRS powder.</li> | <li>MRS agar was prepared by mixing 55g of Difco MRS broth powder and 15g of agar flakes in 1 L of water. The solution was then autoclaved and stored in a 4 degree refrigerator along with the Difco MRS powder.</li> | ||
</div> | </div> | ||
| − | + | <br> | |
| − | + | ||
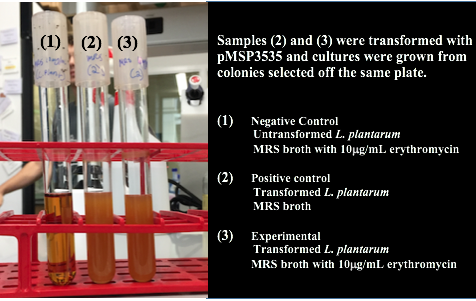
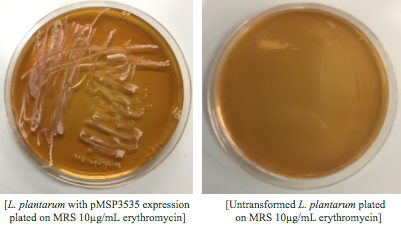
<p style="font-family: verdana">Once we could successfully grow our chosen bacteria, we needed to transform the gram positive, <i> Lactobacillus plantarum</i>, with pMSP3535. In order to do this, we identified and worked with a different protocol than we had ever used in our lab. We attempted several protocols, including Landete 2014 (10) and Speer 2012 (11). However, we found success using a variation of the Welker protocol(12). Welker et al. transformed multiple strains of <i>Lactobacillus casei</i> using varying reagents and yielded different efficiencies between each strain of the species with each variation.</p> | <p style="font-family: verdana">Once we could successfully grow our chosen bacteria, we needed to transform the gram positive, <i> Lactobacillus plantarum</i>, with pMSP3535. In order to do this, we identified and worked with a different protocol than we had ever used in our lab. We attempted several protocols, including Landete 2014 (10) and Speer 2012 (11). However, we found success using a variation of the Welker protocol(12). Welker et al. transformed multiple strains of <i>Lactobacillus casei</i> using varying reagents and yielded different efficiencies between each strain of the species with each variation.</p> | ||
| − | |||
<br> | <br> | ||
<p style="font-family: verdana">The Texas Tech iGEM team helped us by testing the Speer protocol after we had attempted three procedures and hadn't successfully transformed. They were not able to successfully transform using the Speer protocol which suggests that the procedure was either too simplified for researchers who have never transformed gram-positive bacteria, or was not compatible with <i>Lactobacillus plantarum.</i> | <p style="font-family: verdana">The Texas Tech iGEM team helped us by testing the Speer protocol after we had attempted three procedures and hadn't successfully transformed. They were not able to successfully transform using the Speer protocol which suggests that the procedure was either too simplified for researchers who have never transformed gram-positive bacteria, or was not compatible with <i>Lactobacillus plantarum.</i> | ||
Revision as of 06:52, 1 November 2017
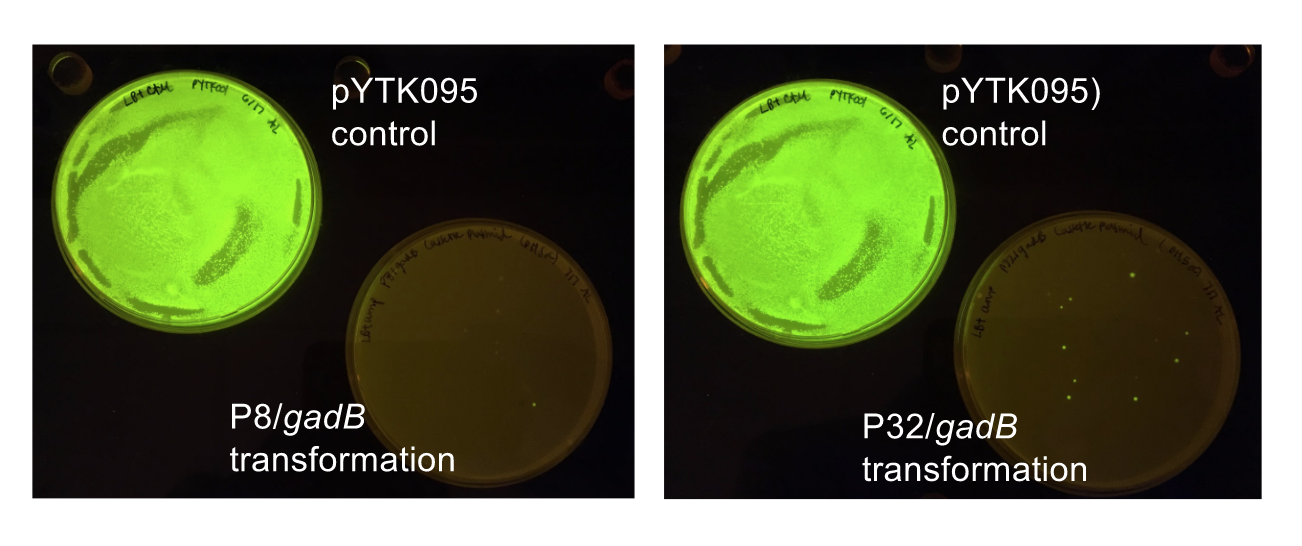
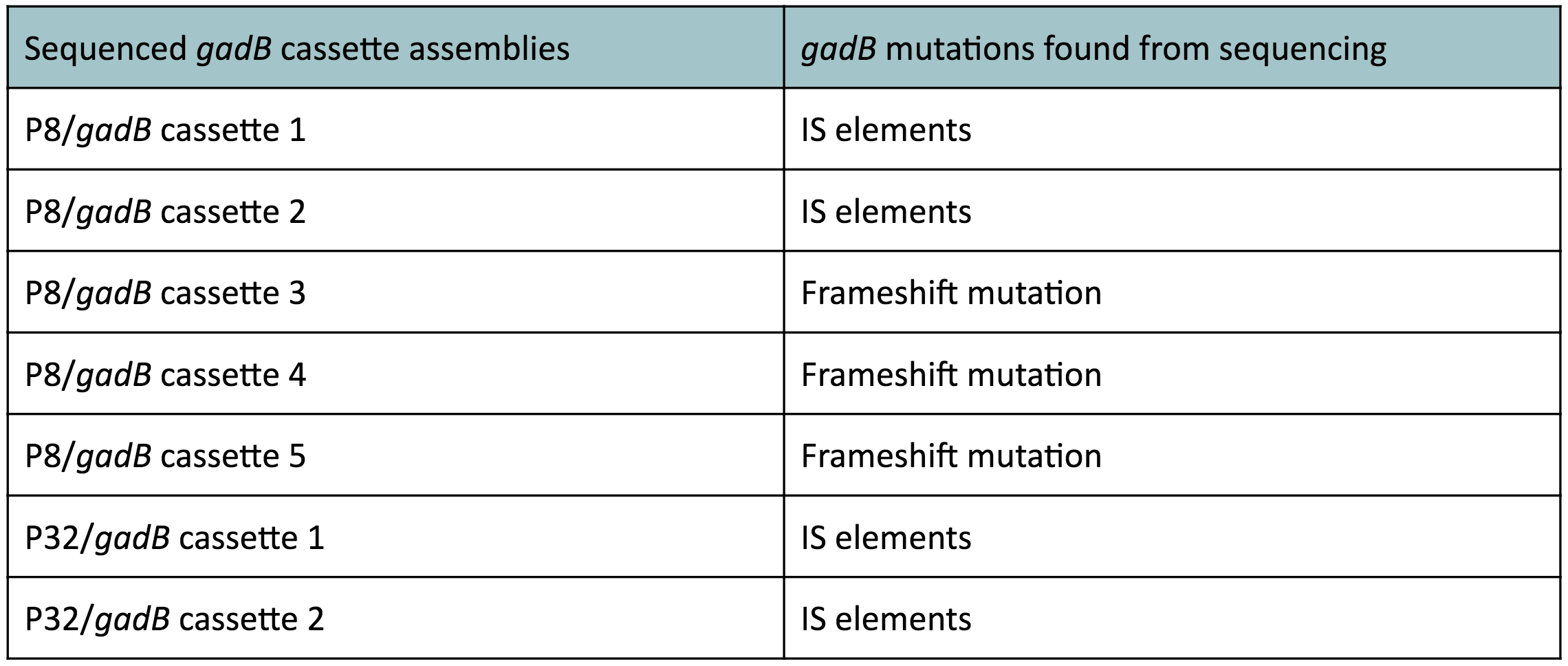
Click on one of the images below to learn more about our results!
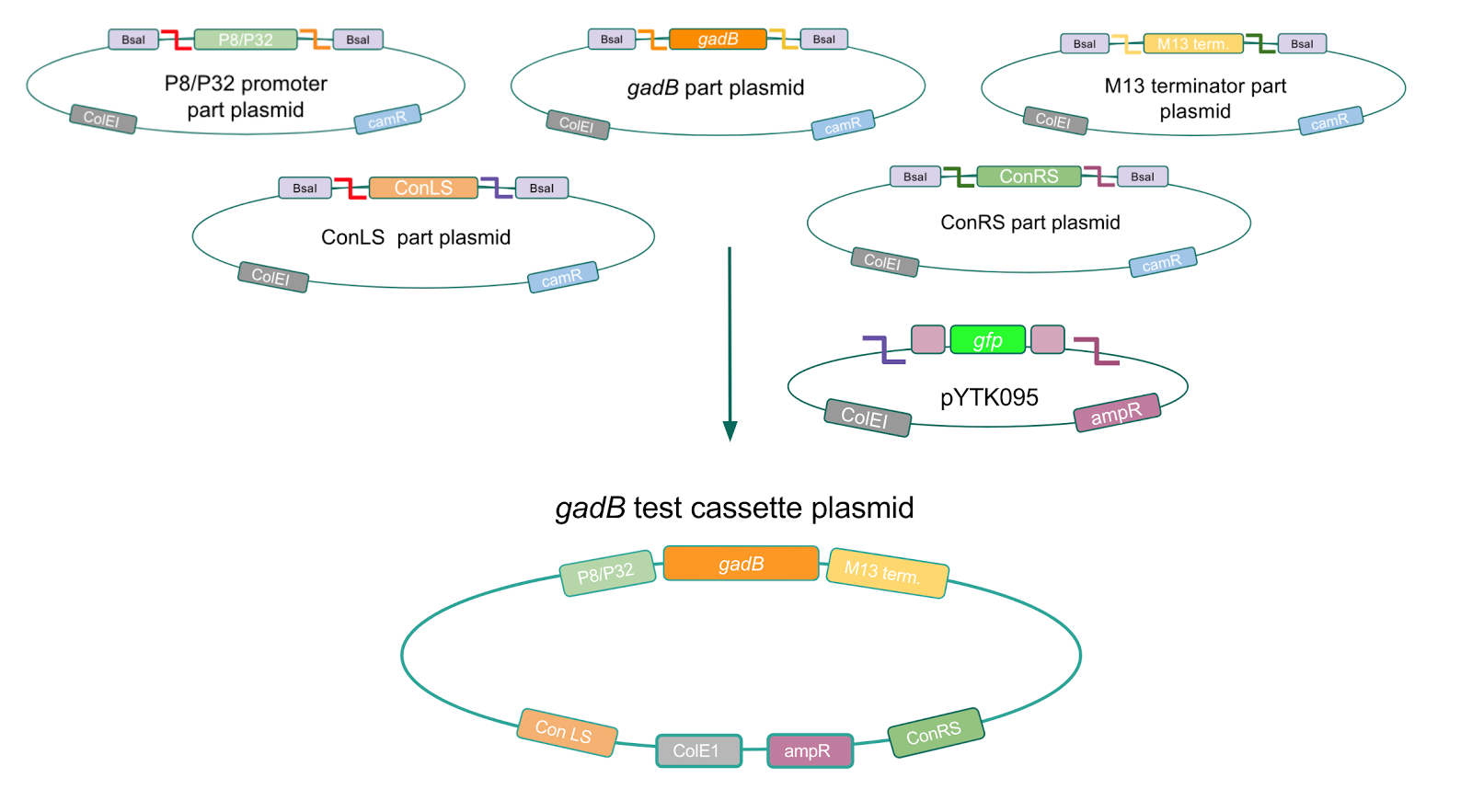
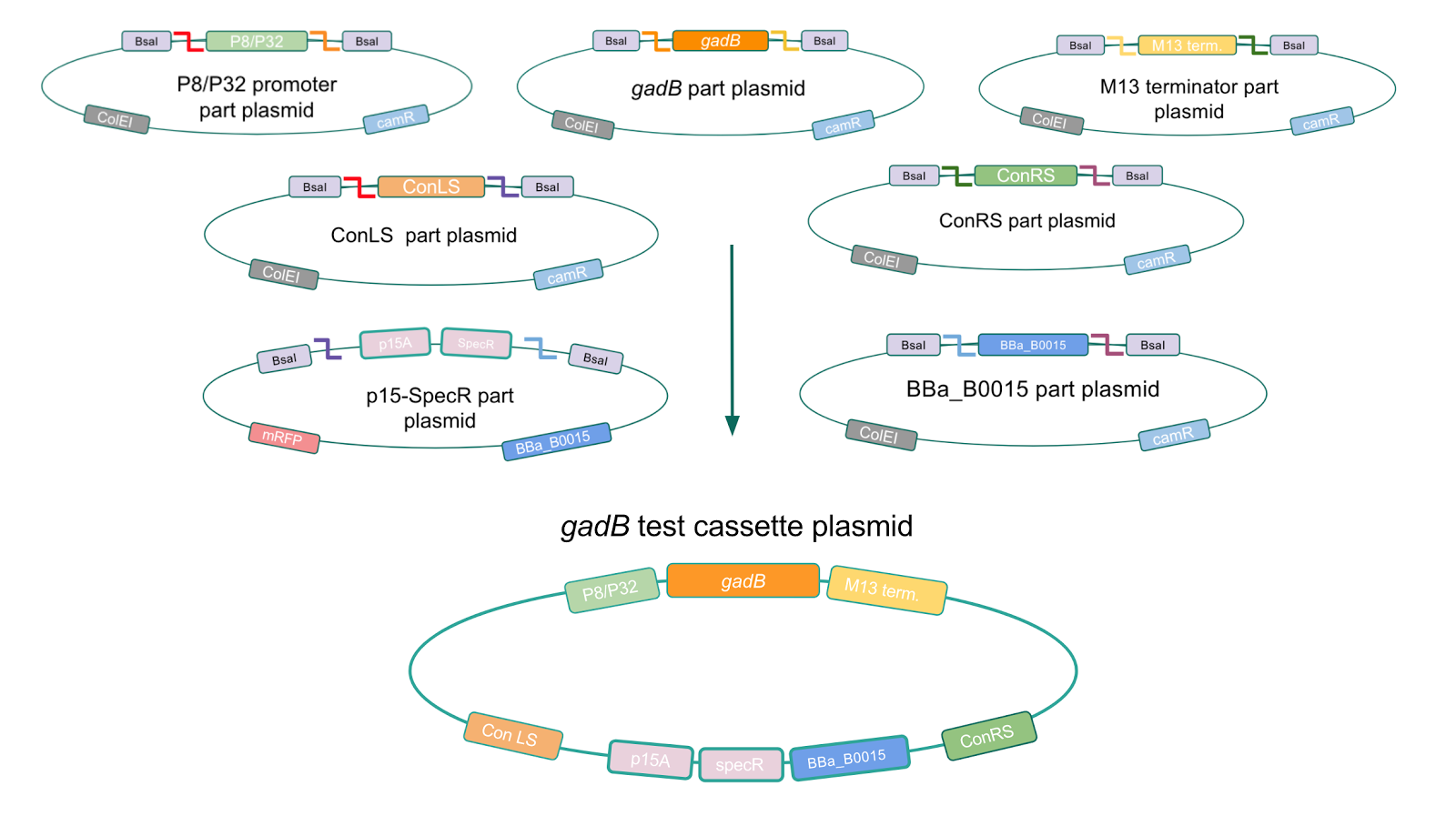
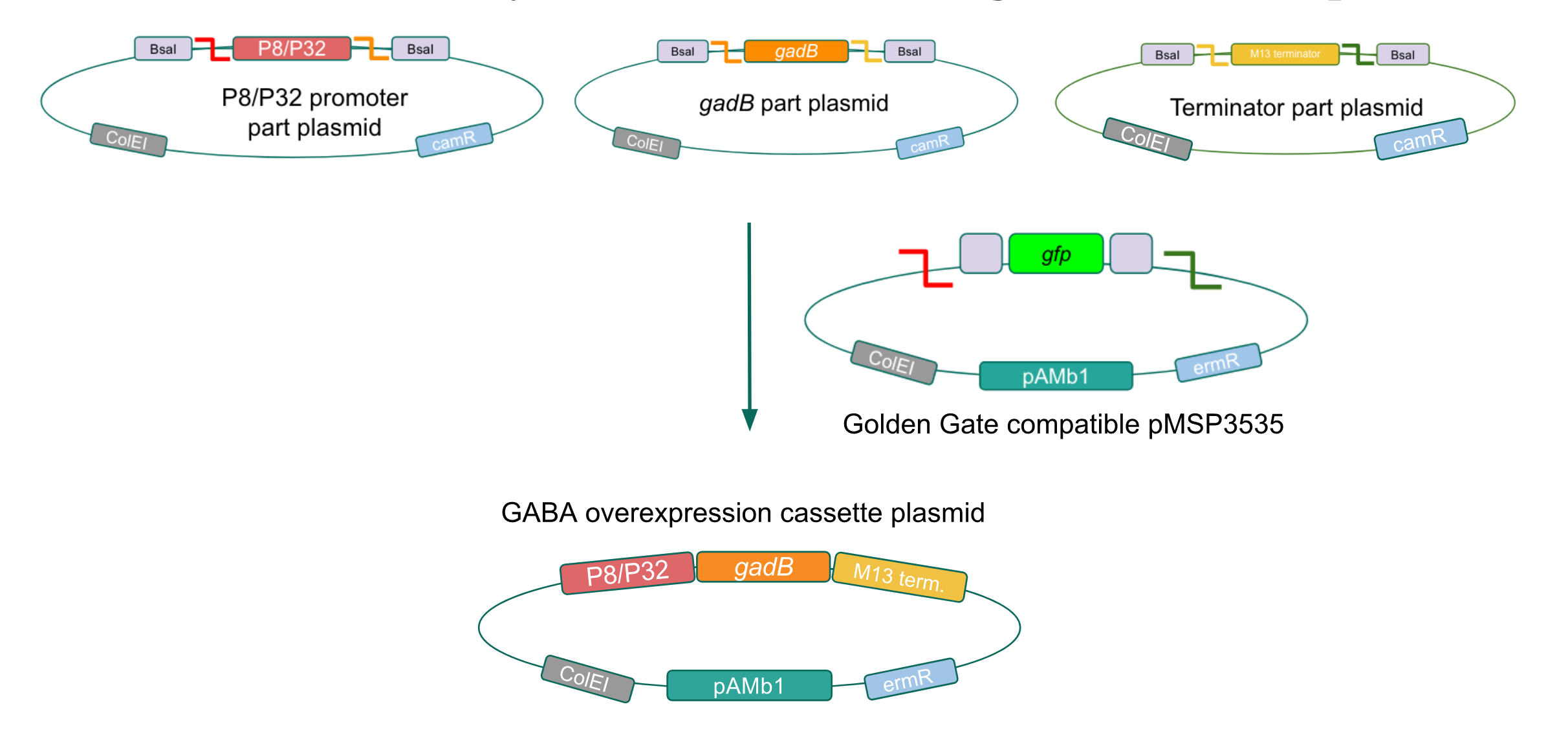
Although bacteria can naturally synthesize GABA, we wanted to increase expression of the gadB gene and subsequently GABA production in order to imbue our probiotic with a more potent medicinal quality, with the idea that this GABA-overproducing probiotic can then be consumed by patients with bowel disorders or anxiety (1). Overexpression of the gadB gene will be accomplished by placing it under the control of either the P8 or P32 constitutive promoters from Lactococcus lactis (2).
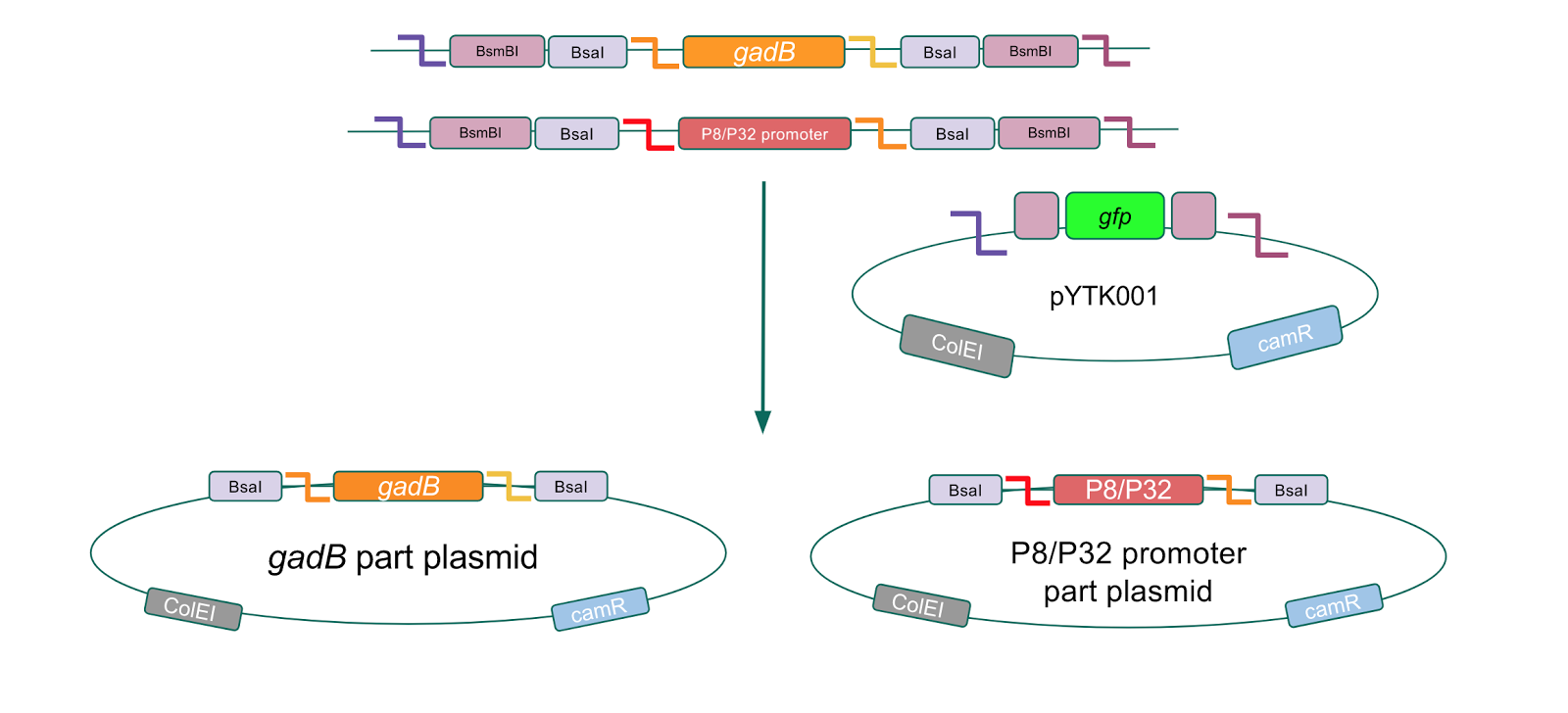
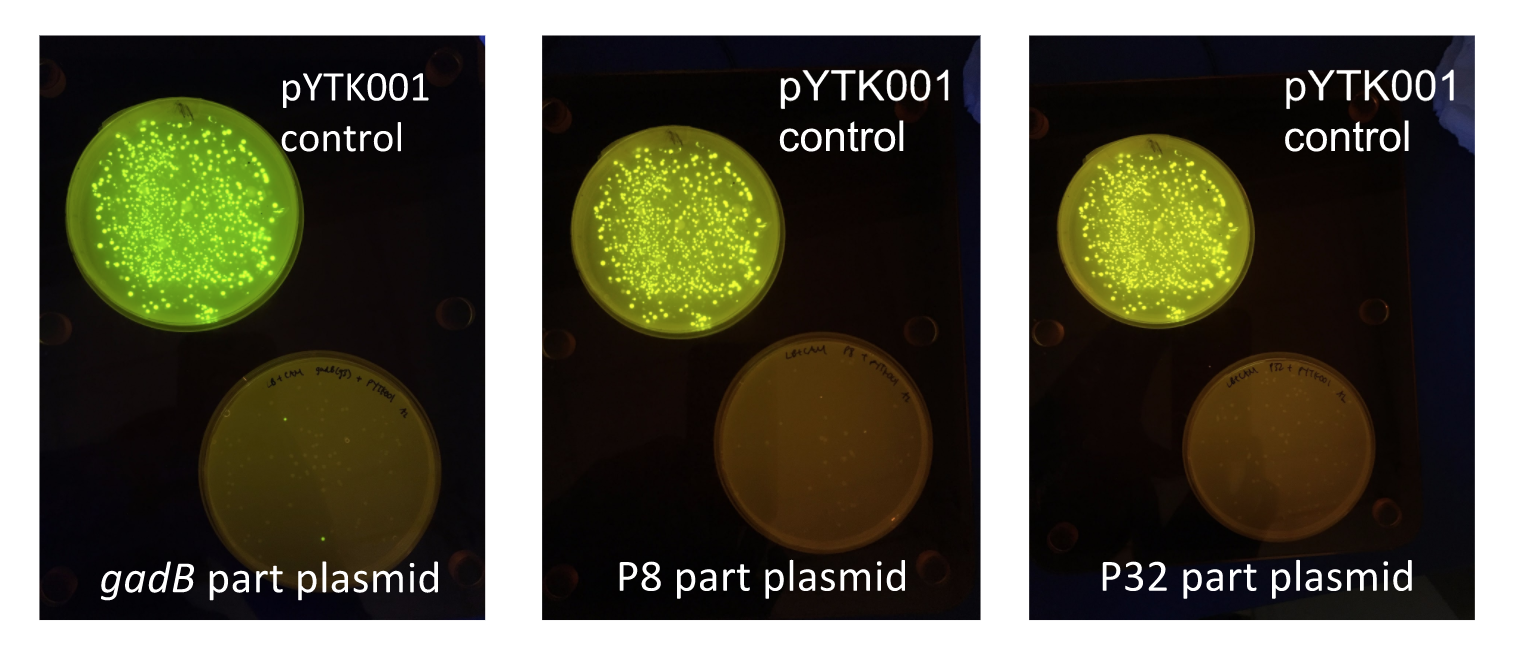
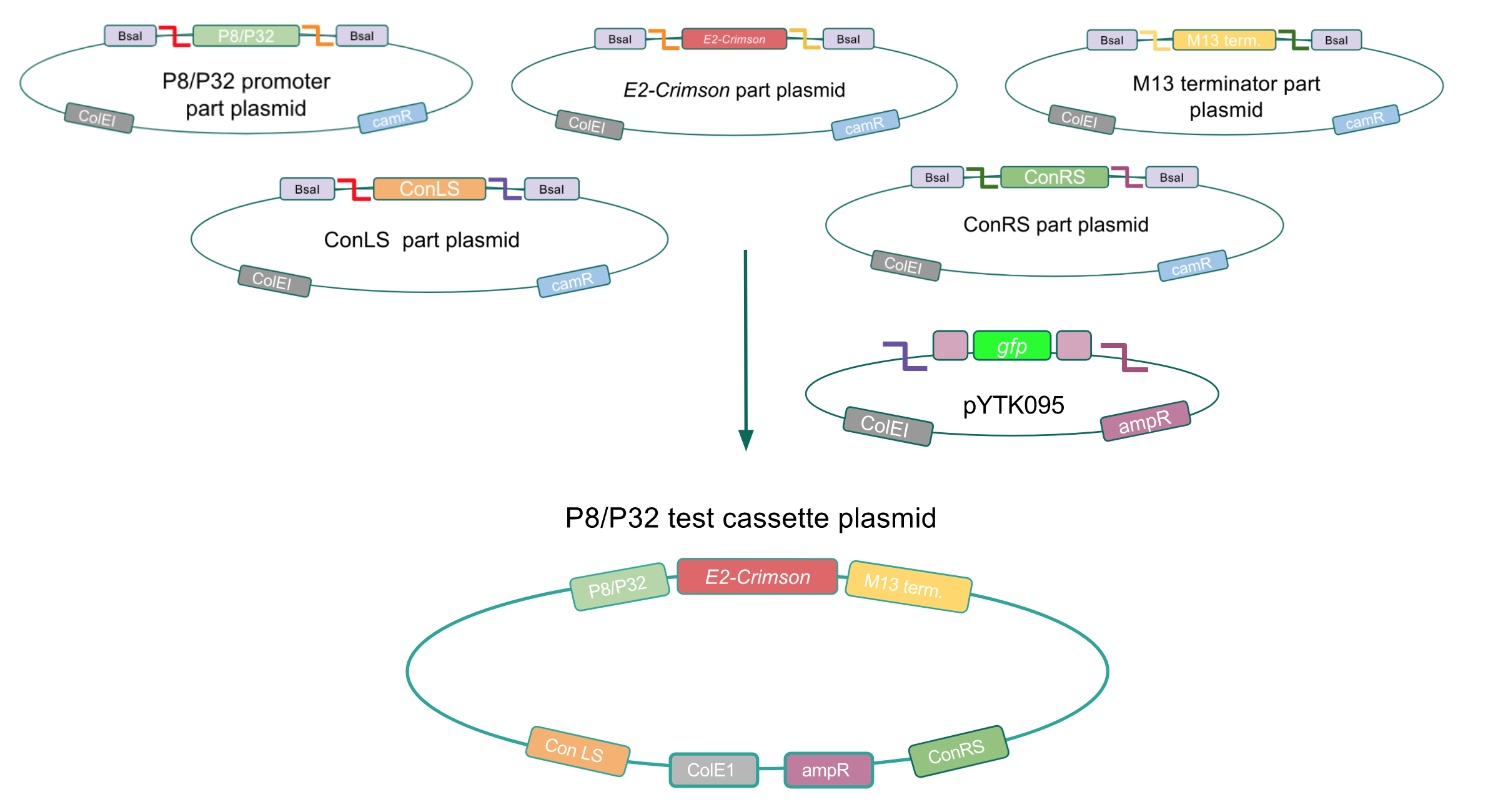
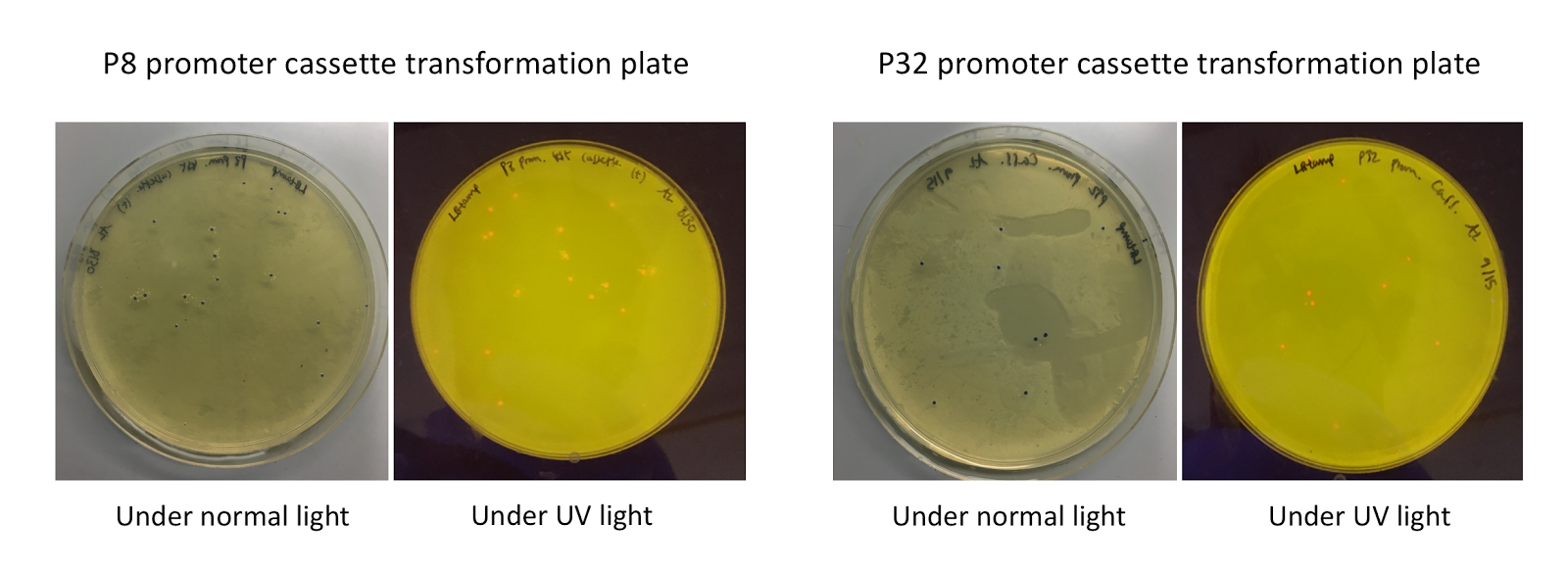
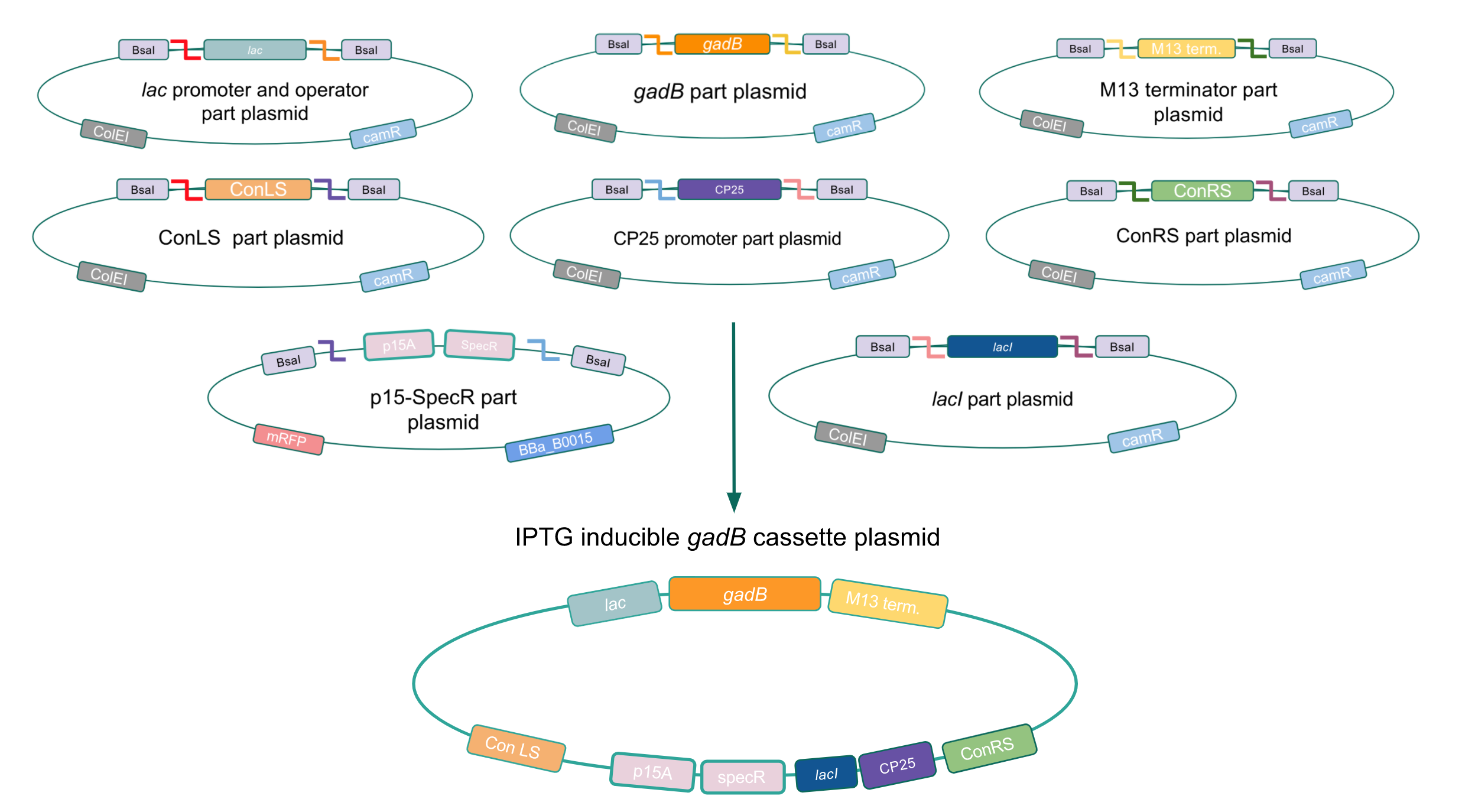
To make our GABA-producing probiotic we first needed to assemble a GABA overexpression cassette plasmid using the Golden Gate assembly method. The intention here is that bacteria containing this GABA overexpression cassette plasmid should produce high levels of GABA. In short, Golden Gate Assembly is a new cloning method that allows for the creation of a multi-part DNA assembly (i.e. cassette plasmid) in a single reaction through the use of DNA parts containing specific, predefined suffixes and prefixes with recognition sites for Type IIs restriction enzymes (e.g. BsmBI and BsaI). The specificity of these suffixes and prefixes provides directionality of the desired DNA parts during the assembly process. For our purposes, we used the MoClo Yeast Tool Kit developed by John Dueber (3).
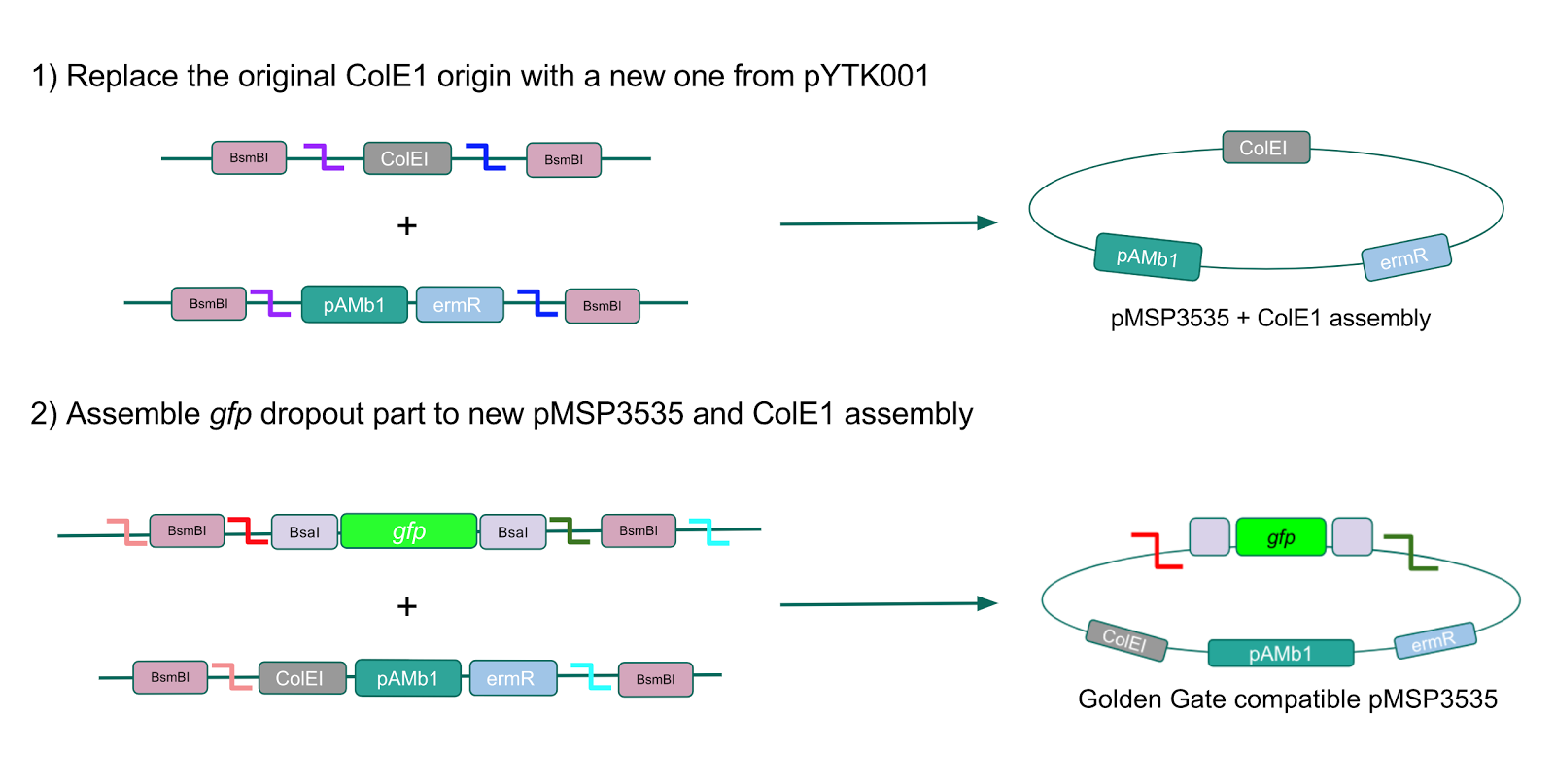
Creating a Golden Gate compatible shuttle vector
We wanted to assemble our final GABA overexpression cassette plasmid using the shuttle vector pMSP3535 as the backbone (Fig. 9). To do this, we first needed to make pMSP3535 Golden Gate compatible (i.e. free of BsaI restriction sites and containing correct overhangs for cassette assembly). We chose to work with pMSP3535 as it contains both a ColE1 origin for replication in E. coli and a pAMb1 origin for replication in Gram-positive bacteria including Lactobacillus species (6). Additionally, the pMSP3535 vector contains the resistance gene for erythromycin, of which Lactobacillus plantarum is naturally susceptible (7).
The process of making the pMSP3535 vector Golden Gate compatible involved two steps: 1) assembling the pMSP3535 backbone (pAMb1 origin and erythromycin resistance gene) with a new ColE1 origin; 2) assembling a gfp dropout part to the assembly of the pMSP3535 backbone and the new ColE1 origin (Fig. 10).
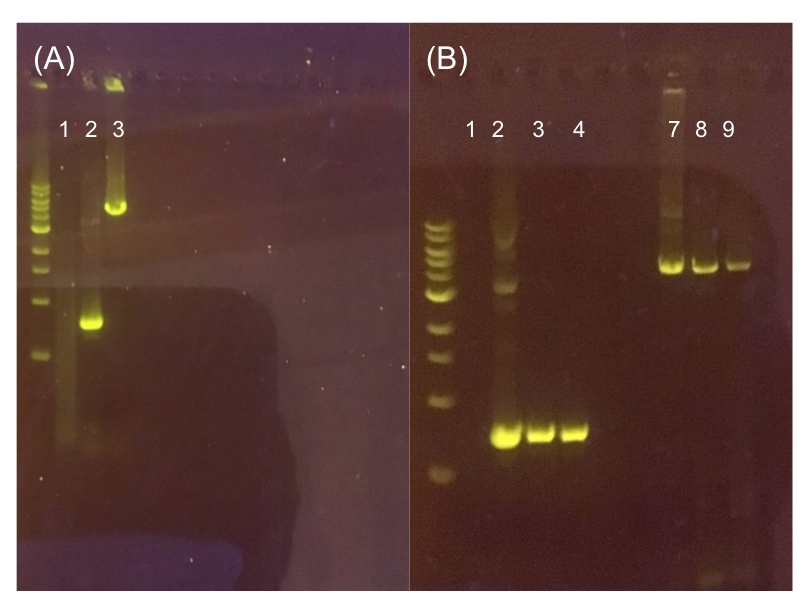
Assessing erythromycin susceptibility of E. coli
Because we are creating our Golden Gate compatible pMSP3535 shuttle vector in E. coli, we wanted to determine the natural susceptibility of E. coli to erythromycin as the minimum concentration to use has not been established clearly in the literature. Thus, we performed an erythromycin minimum inhibitory concentration test in liquid LB media (Fig. 13). After one-day incubation, we observed that E. coli was resistant up to around 150 µg/mL of erythromycin. From this experiment, we have determined that the optimal erythromycin concentration for selecting against E. coli in liquid culture is around 200-250 µg/mL.