Microfluidics
Create for wisdom of Life
1 The Selection Chip
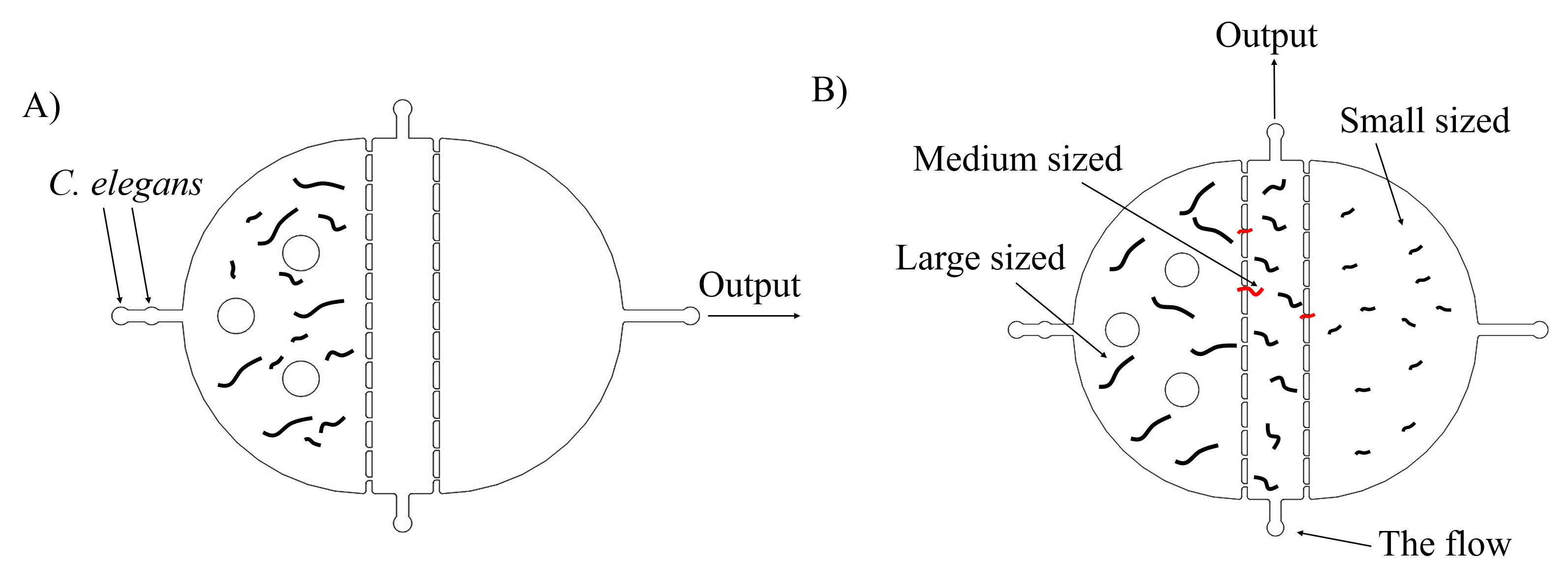
Most researches in Caenorhabditis elegans show that inserted genes can express well in worms at L4 stages. Thus, we need to select appropriate stages of worms to get the best experimental results. The simple method is to distinguish them by sizes. Worms in L4 stage have medium sizes. There are two plans of selecting worms. The first one is using microfluidics. After the flow with worms go through this chip, the medium sized worms can remain in the medium chamber. And get them by injecting the flow from the bottom entry to the top exist. (Fig. 1)
The second plan is do the synchronization of worms, which is utilized to get a large number of worms at the same stage (链接到微流控protocol). After getting embryos (Fig. 2) by bleaching adults, we culture them and can get a large number of worms at the same stages after three days. our synchronous rate is
\frac{(the\,number\,of\, the\,worms\,at\,L4)*100}{the\, number\, of\, all\, worms}
The successful rate can reach to about 80%.
Compared with these two methods, we think the synchronization method is better. You can get more detail results by clicking here. (https://2017.igem.org/Team:SUSTech_Shenzhen/Results)
2 The Gaussian Chip
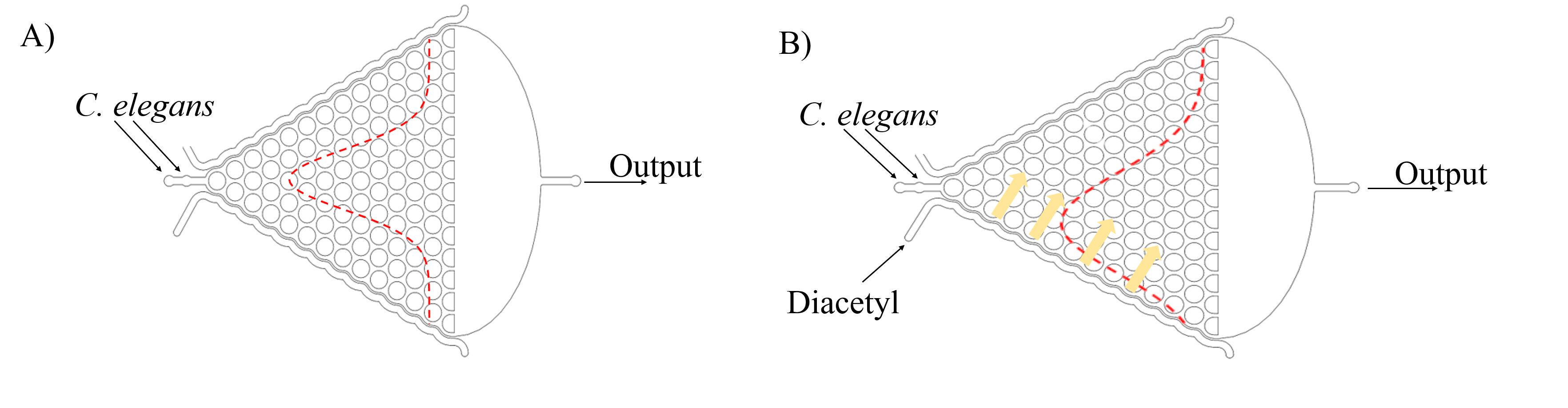
In order to study locomotion of C. elegans populations, we design the Gaussian Chip, a pillar-filled area, where the pillars are designed such that they allow crawling-like behaviors even though worms are immersed in liquid environment[2].(Fig. 3) [1].
After deciding to use this microfluidics, we are inspired that it is like the Galton board[3].[2]. (Fig. 4)
We assume that the probability for C. elegans choose to go left or right is equal when it passes a crossing. (Fig.5)
And then we can assume that C. elegans is similar to balls in the Galton board. The force of slow buffer flow acting on worms is like gravity acting on balls. Both of the distribution is Gaussian distribution. Given that we need to make sure whether our insertions will affect the olfactory receptor neuron pair after C. elegans being injected the target genes, we injected diacetyl (2-nonanone) that C. elegans prefers (repulse) into the right (left) channel to make a concentration gradient on the Gaussian chip. Because of the gradient, worms tend to the side filled with diacetyl, causing Gaussian distribution changed. (Fig. 6)
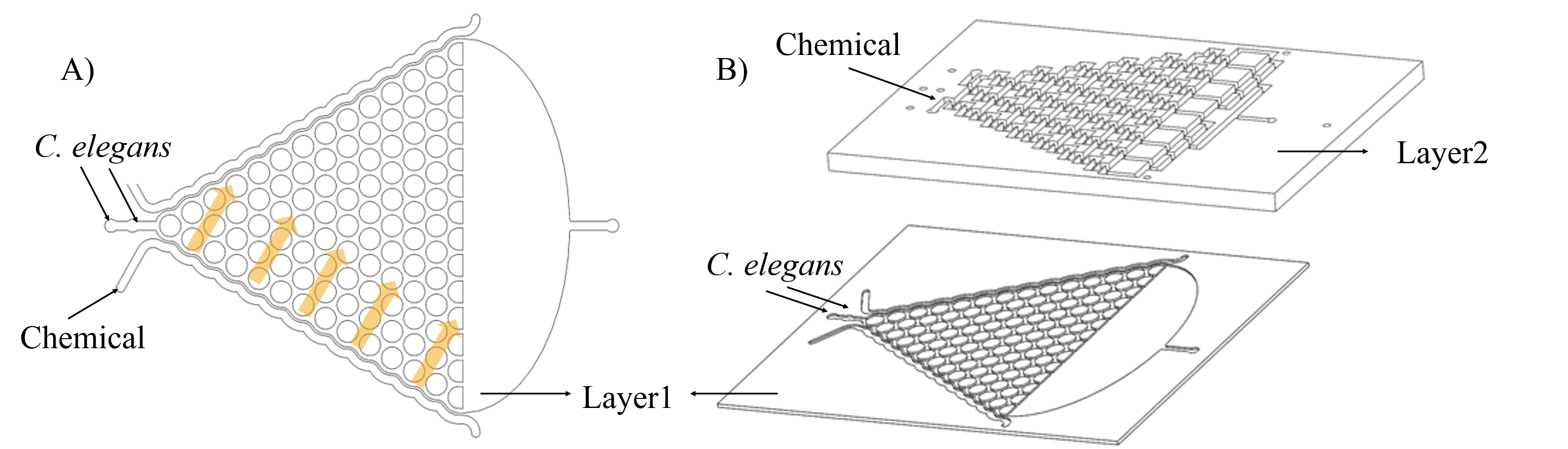
In order to make a concentration gradient, we come up with two methods to get it. (Fig. 7)
In order to simulate the process of diffusion, we make a diffusion model to guide us. More details.(https://2017.igem.org/Team:SUSTech_Shenzhen/Chemical_Diffusion_Model)
Both of these methods can be carried out in theory. But in experiment, we find the method 1 (Inject chemicals into the side of layer 1) is better.
3 The Immobilization Chip
After studying worms’ group behaviors and proving their olfactory neurons are not being affected by exogenous gene, we could study their individual neuron activity and behavioral response. Traditionally, anesthetics and glues are utilized to immobilize worms. However worms will be damaged in these and it will create difficulties in studying its behavioral response. Thus, we designed two kinds of microfluidic chips to allow high-resolution microscopic imaging on chip without damaging for worms. (Fig. 8)
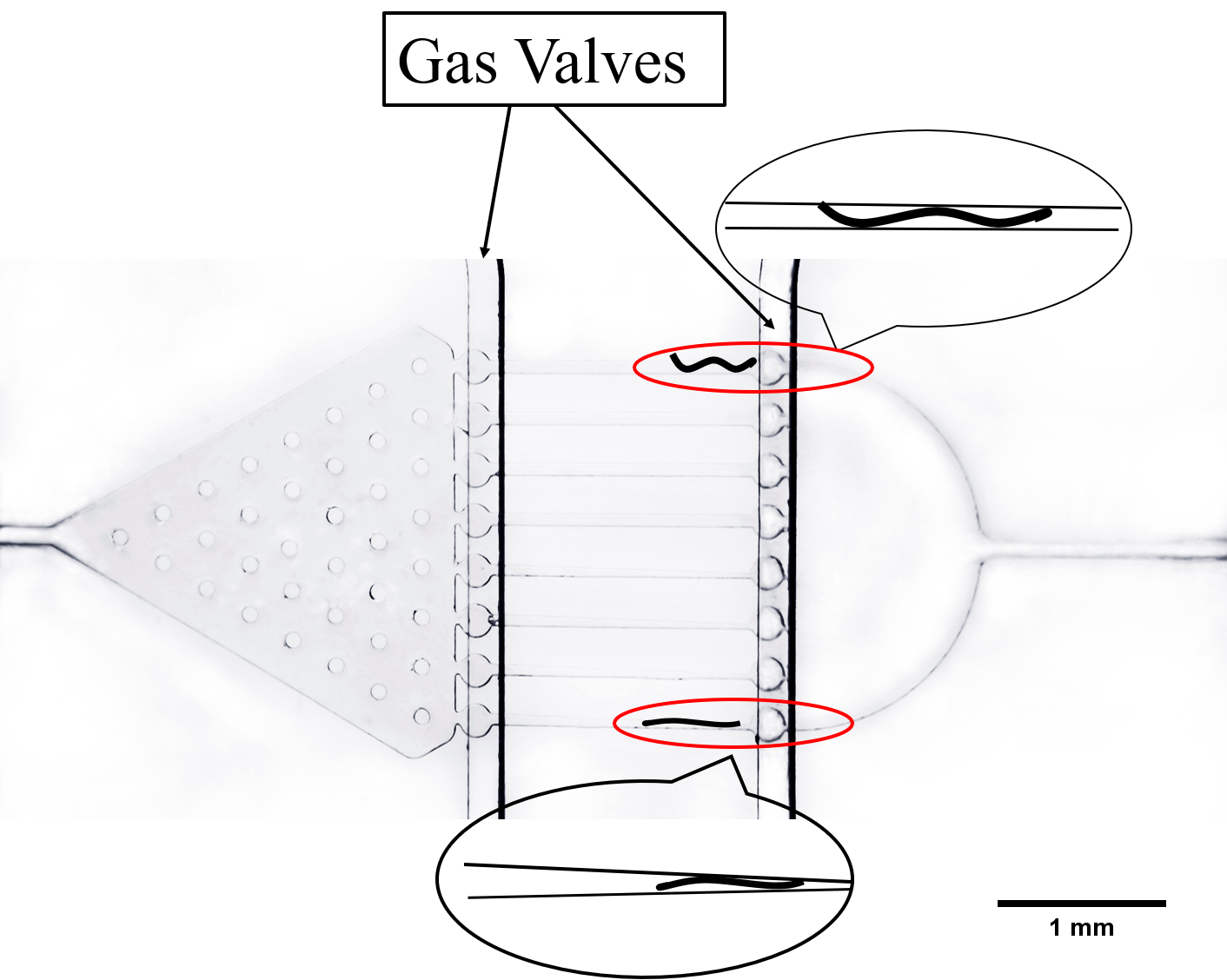
The first one is used to research the imaging of neuronal response, called worm clamps or traps[3].Worms are trapped in the wedge-shaped channel. Trapped worms can then be released by reversing the flow. Traps allow worms’ movement in x-y plane but restrict it in z direction, and it is also utilized to study the contraction and elongation of their heads and research the neuronal activity by detecting calcium indicator GEM-GECO in imaging software.
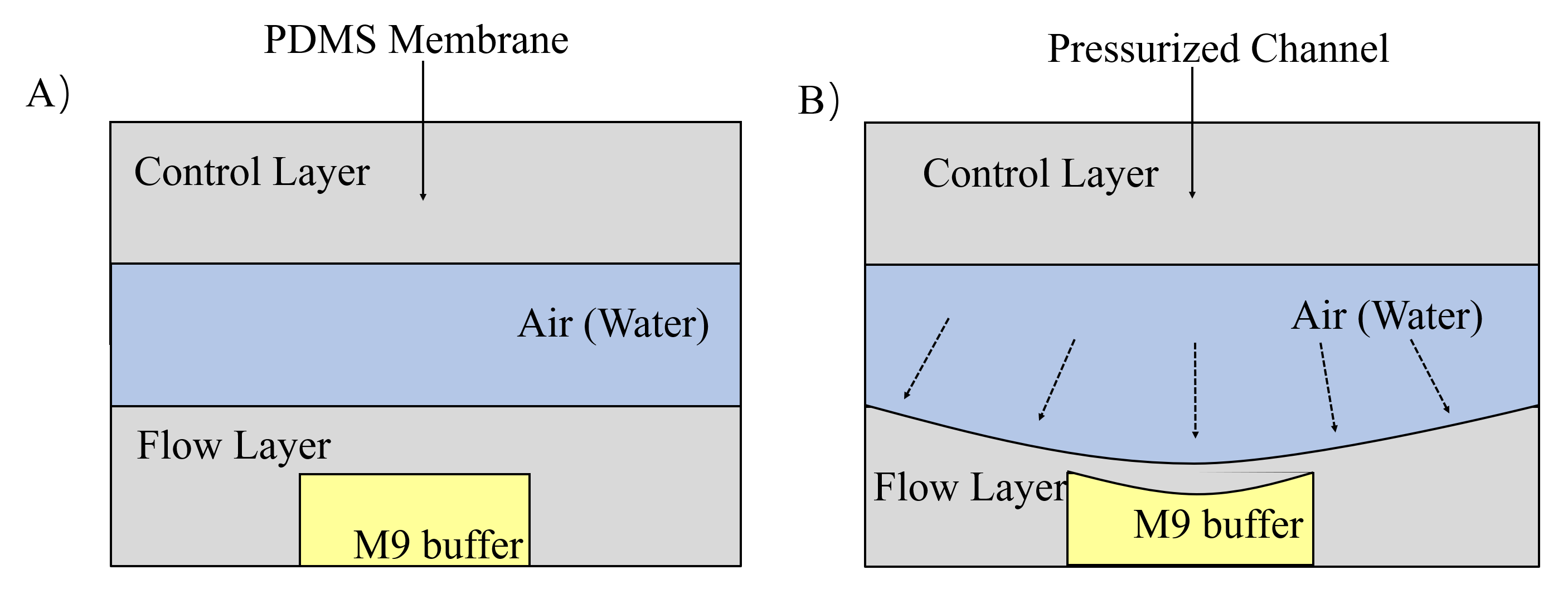
The next channel is compressed and rectangular, called parallel channel. In order to immobilize worms in this channel, we design two gas valves to block the exit and entry by compressing PDMS (a flexible and Easily deformed material) under the pressure made by water or air. (Fig. 9) [4]
The contents above are our all hardware parts in microfluidics. You can learn more details in results. (https://2017.igem.org/Team:SUSTech_Shenzhen/Results)
References
- ↑ Albrecht, D.R., and Bargmann, C.I. (2011). High-content behavioral analysis of Caenorhabditis elegans in precise spatiotemporal chemical environments. Nat. Methods 8, 599-605.
- ↑ Bean machine. (2017, October 5). In Wikipedia, The Free Encyclopedia. Retrieved 12:46, October 22, 2017, from https://en.wikipedia.org/w/index.php?title=Bean_machine&oldid=803992086
- ↑ Hulme, S. E., Shevkoplyas, S. S., Apfeld, J., Fontana, W., & Whitesides, G. M. (2007). A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab on A Chip, 7(11), 1515.
- ↑ Unger, M.A., Chou, H.P., Thorsen, T., Scherer, A., and Quake, S.R. (2000). Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113-116.