Testing constitutive Lactococcal promoters in E. coli
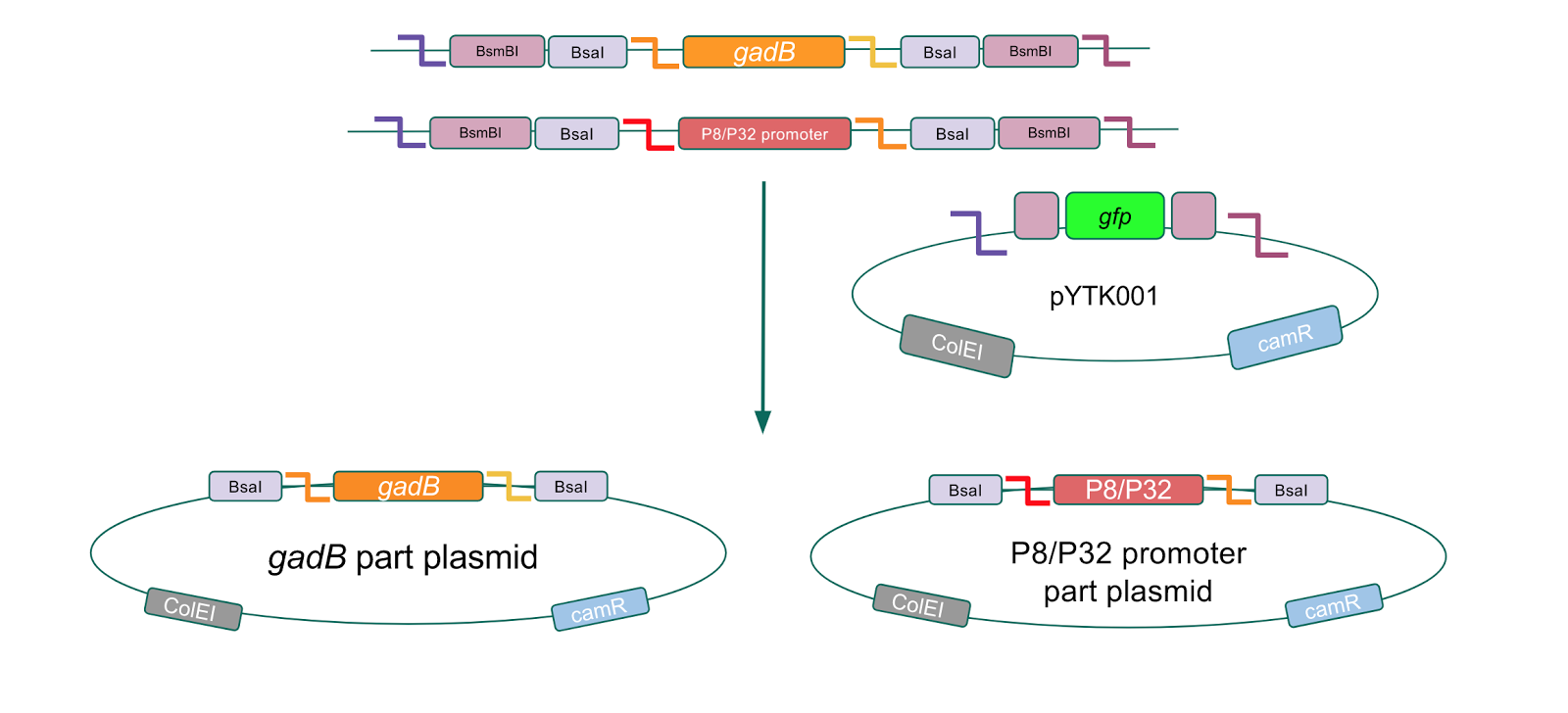
After successfully creating the gadB gene and P8/P32 promoter part plasmids, the functionality of these part plasmids were then assessed by assembling them into cassette plasmids.
Although the functionality of the Lactococcal lactis P8 and P32 constitutive promoters has been well characterized in Gram-positive bacteria such as Lactobacillus, we wanted to test if these promoters could function well within E. coli. To do this, we created test cassette plasmids containing the E2-Crimson reporter gene (which encodes a red fluorescent protein) inserted downstream of either the P8 or P32 promoters using BsaI Golden Gate assembly. To create these test cassettes, we used the P8/P32 promoter part plasmids, the E2-Crimson part plasmid, an M13 terminator part plasmid, connector part plasmids, and the pYTK095 vector as the backbone (Fig. 3). If the P8 and P32 promoters are functional in E. coli, we expected to observe red fluorescence in colonies transformed with our test cassette plasmids.
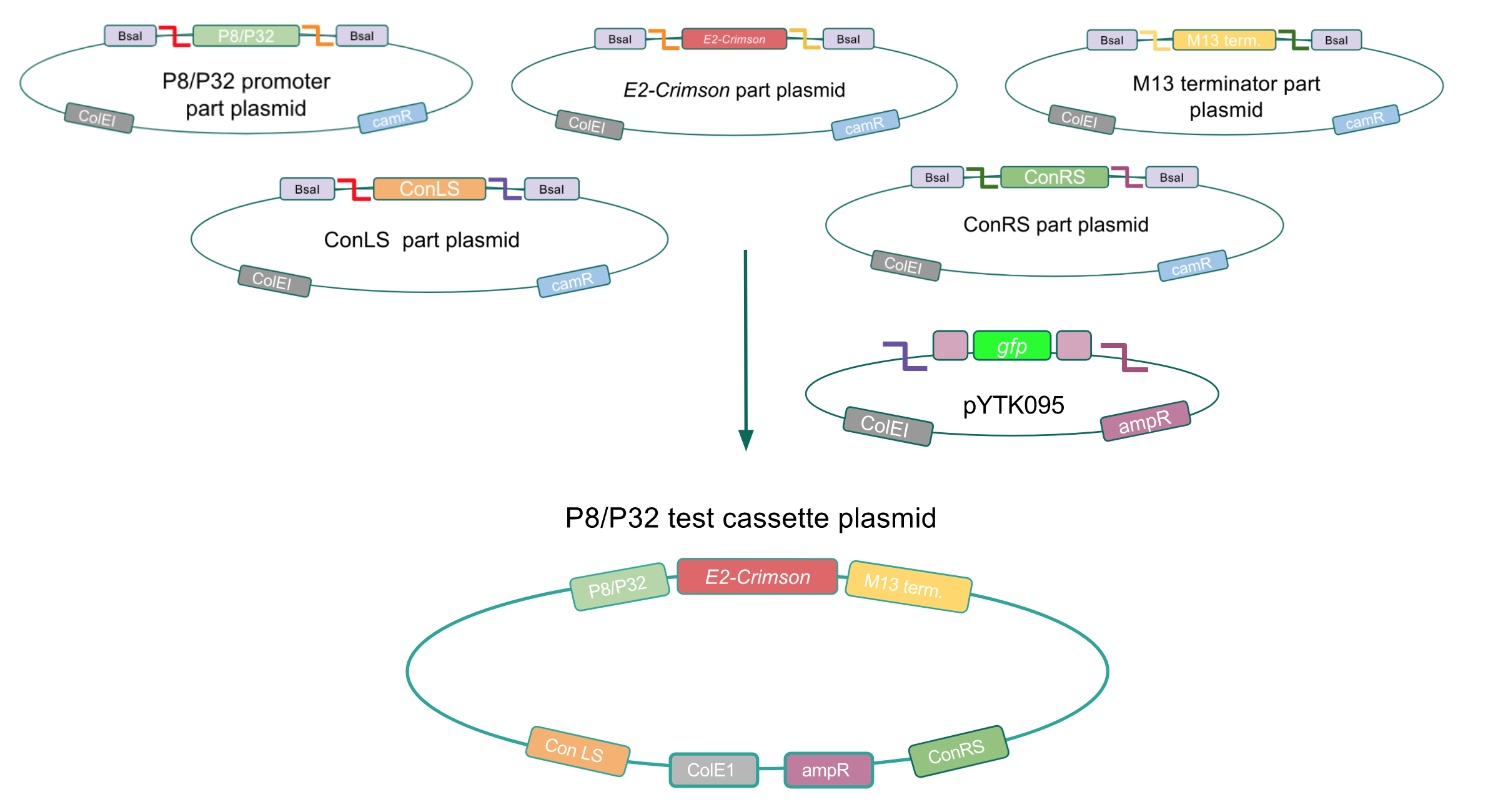
Figure 3. Golden Gate assembly process of the P8 and P32 test cassette plasmids.
Upon first look, E. coli colonies transformed with these assemblies appeared purple-blue in color. This phenotype was due to the expression of the red fluorescent protein. Further, we noticed that E. coli colonies transformed with these assemblies fluoresced red under UV light, indicating that the P8 and P32 promoters are indeed expressing the E2-Crimson reporter gene and thus are functional in E. coli (Fig. 4).
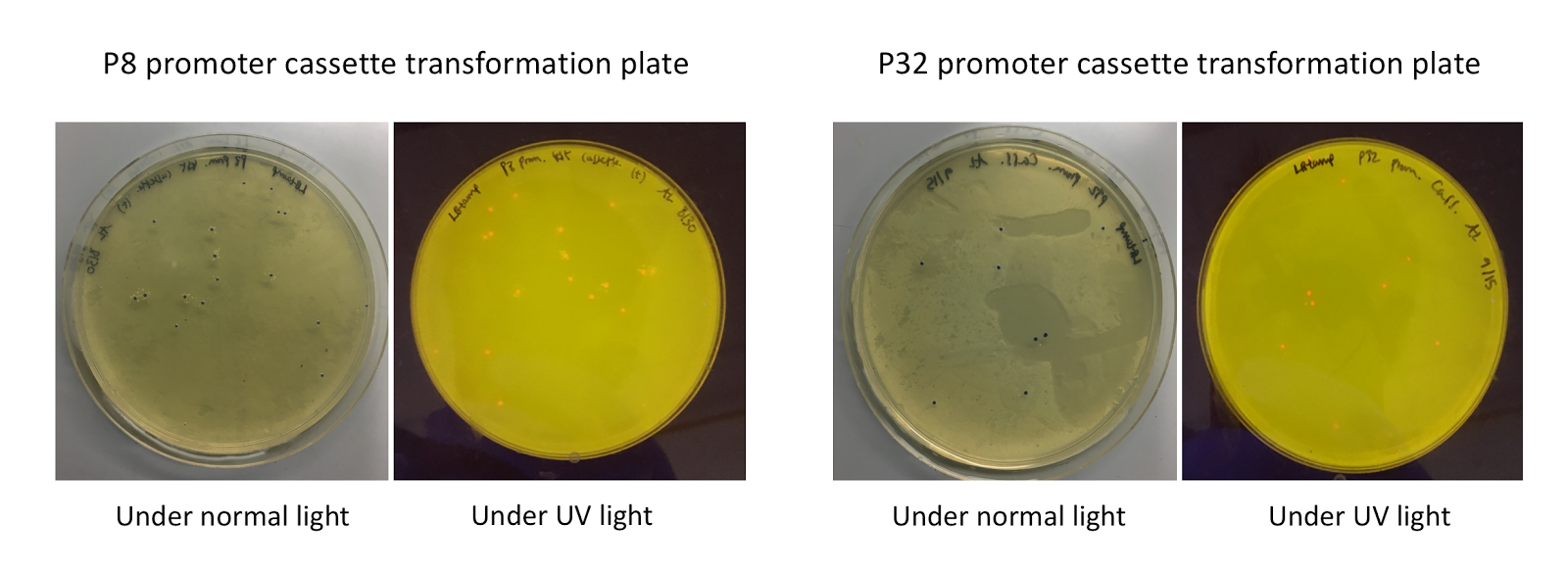
Figure 4. P8/P32 test cassette plasmid transformation plates, under normal and UV lights. Under normal lights, the colonies appeared purple-blue in color. Under UV, the colonies fluoresced red.
Back to Top
Testing gadB overexpression in E. coli
Using Golden Gate Assembly, we created cassette plasmids to test if the gadB gene could be overexpressed in E. coli via the P8 and P32 promoters. These cassette plasmids contained the gadB gene inserted downstream of either the P8 or P32 promoters. To make our gadB test cassette plasmids, we used pYTK095 as our backbone and part plasmids containing the P8/P32 promoters, gadB gene, M13 terminator, and connector sequences (Fig. 5).
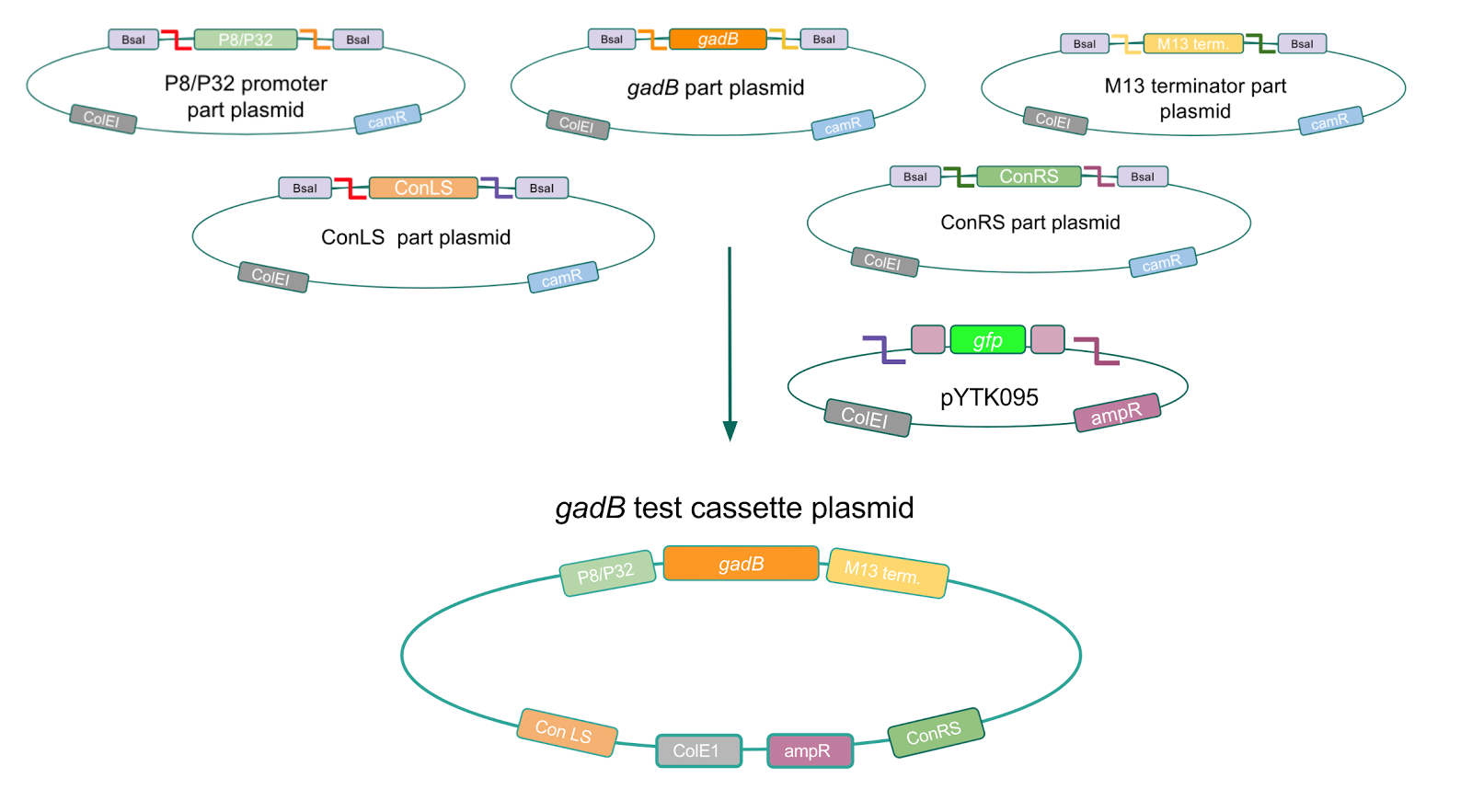
Figure 5. Golden Gate assembly process of P8/
gadB and P32/
gadB test cassette plasmids.
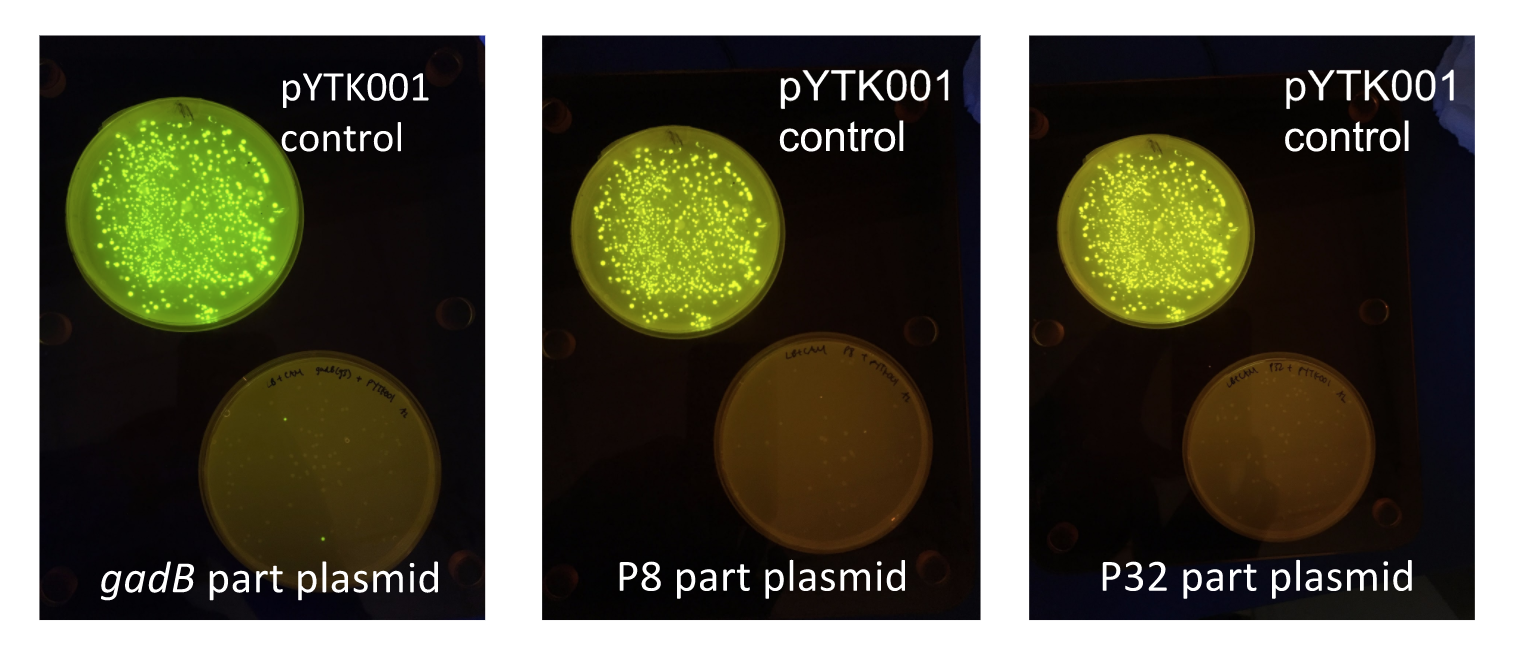
Similar to the pYTK001 entry vector in part assembly, the pYTK095 backbone used for cassette assembly contained a gfp reporter gene that is replaced by sequences of interest. This allowed us to easily perform a phenotypic screen for positive colonies. Non-fluorescent colonies may potentially have had the correct cassette assembly, while fluorescent colonies did not (Fig. 6).
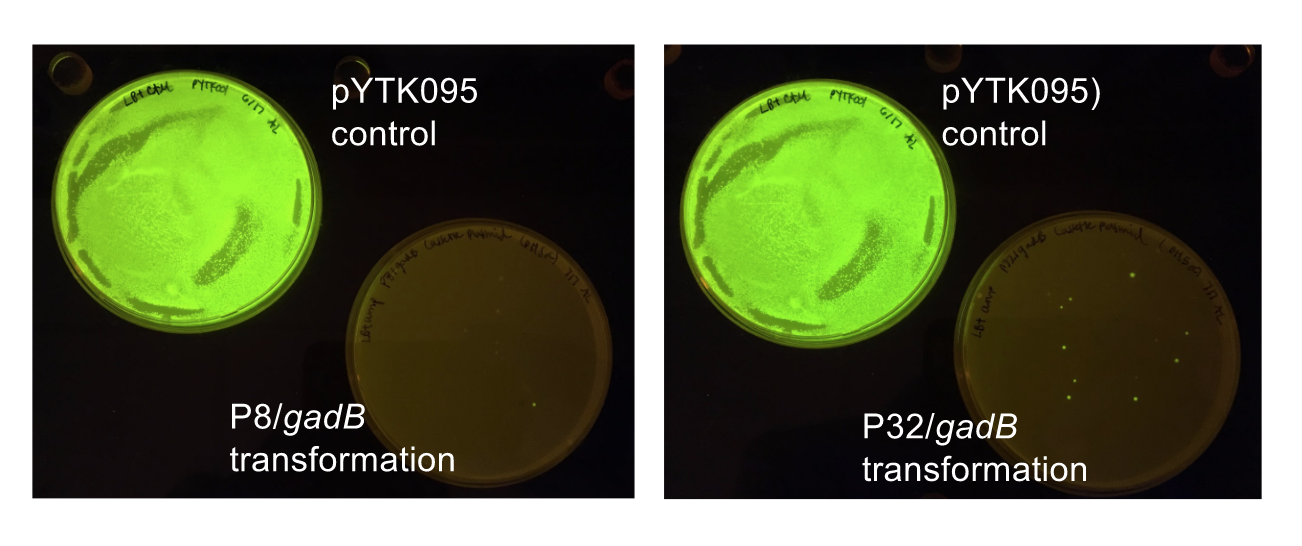
Figure 6. P8/
gadB and P32/
gadB cassette E. coli transformations alongside a pYTK095 control transformation under UV illumination. Potentially positive colonies containing the correct assemblies appeared non-fluorescent, while negative colonies appeared fluorescent.
The non-fluorescent colonies were then screened using colony PCR, and positive colonies were then miniprepped and sequenced. The sequencing results indicated that there were several mutations within the gadB gene in the samples. These mutations are recorded in Table 1.
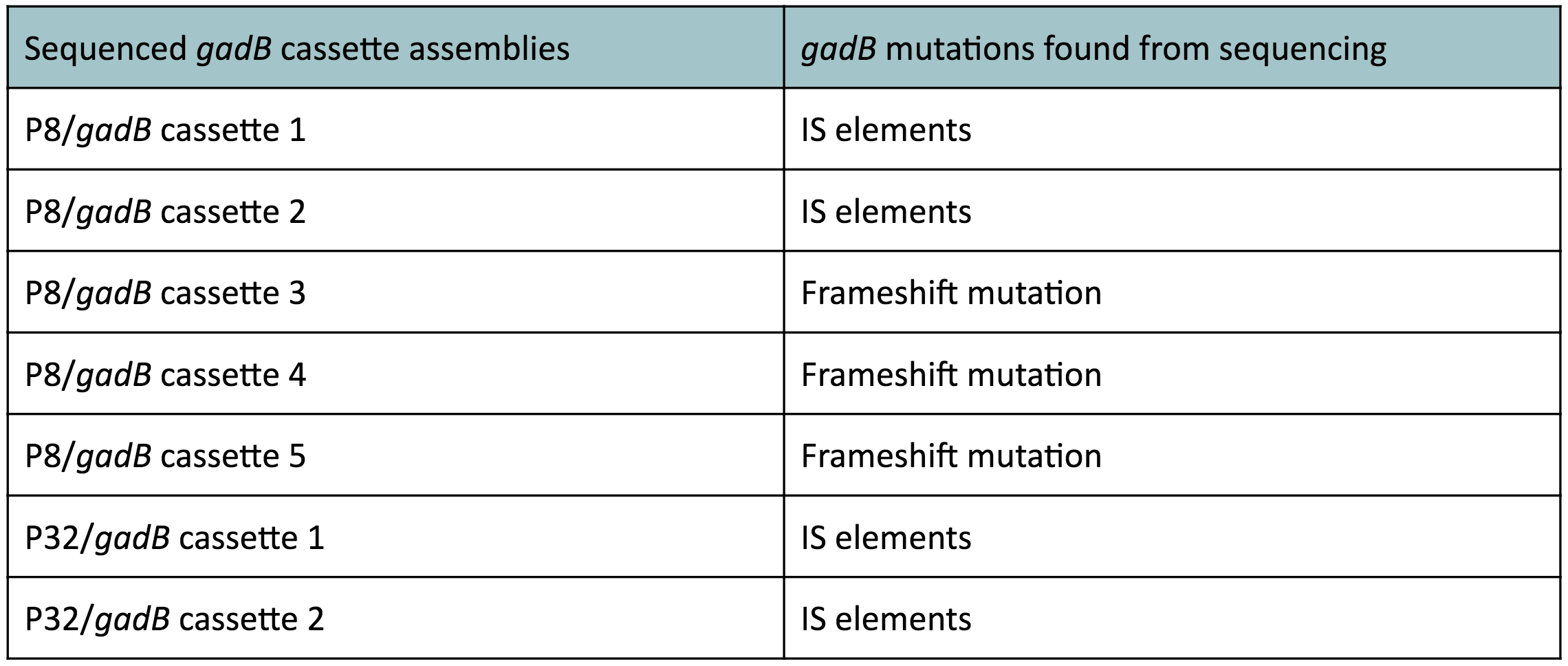
Table 1. gadB mutations in sequenced P8/
gadB and P32/
gadB overexpression cassette plasmids.
Along with being one of the canonical amino acids utilized in protein synthesis, glutamate plays an important role as the main amino-group donor in the biosynthesis of nitrogen-containing compounds such as amino acids and nucleotides (4, 5). Thus, we hypothesized that gadB overexpression via the P8 and P32 constitutive promoters and the high-copy-number ColE1 origin induced a high metabolic load on the cells by shunting away glutamate from essential anabolic pathways. Additionally, having high gadB expression does not confer a selective advantage to the cells. We believed that transformants containing the mutationally degraded gadB gene were favored in the population, as "breaking" the metabolically-taxing gadB gene gave these transformants a competitive advantage, allowing them to utilize glutamate sources towards growth. In contrast, transformants containing the functional gadB gene were selected against due to having a depletion of glutamate needed for important cellular processes.
To troubleshoot this problem, we decided to use a backbone containing the low-copy number p15A origin for the cassette assembly (Fig. 7).
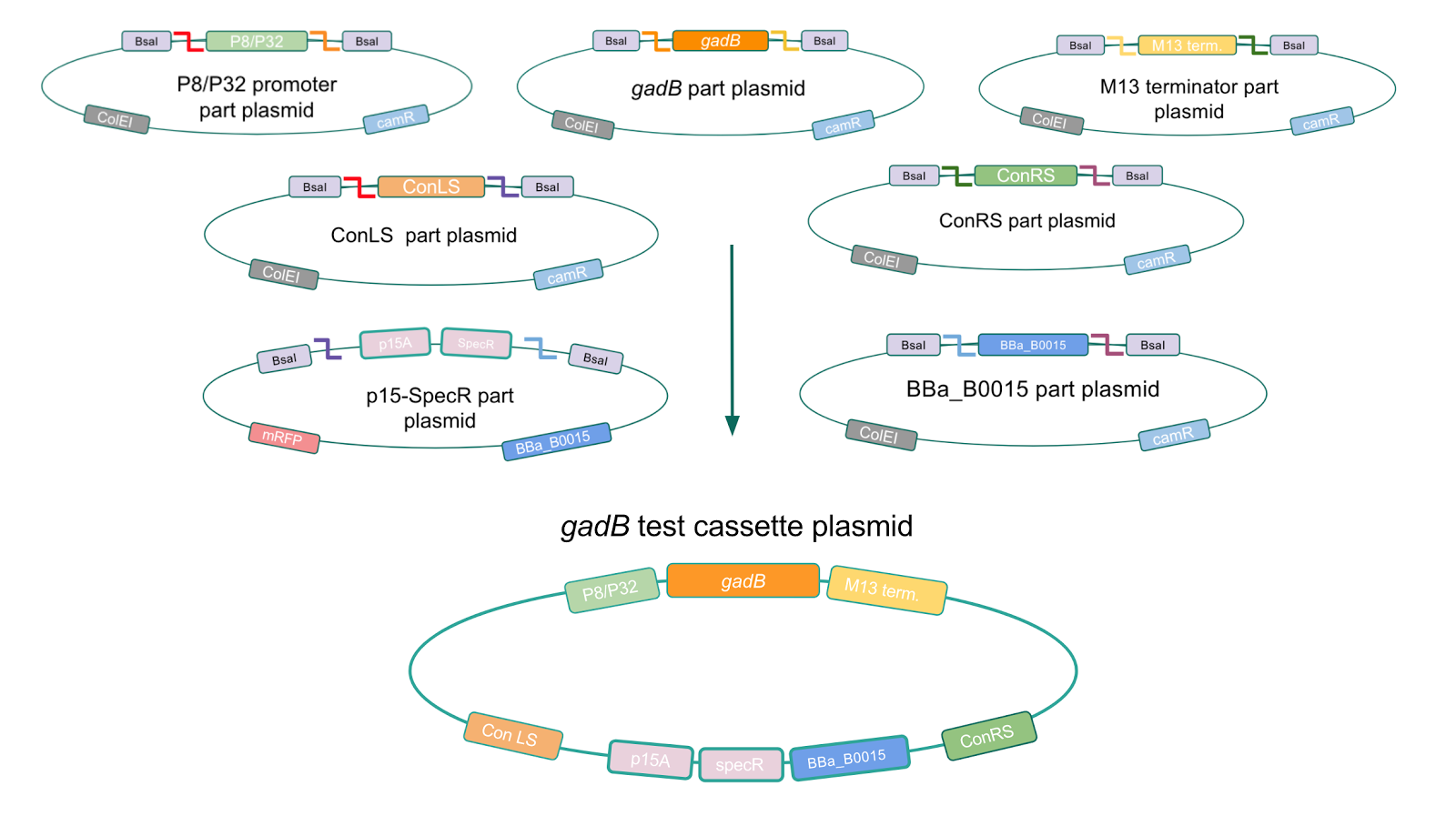
Figure 7. Golden Gate assembly process of P8/
gadB and P32/
gadB test cassette plasmids using a backbone containing the low-copy number p15A origin.
Copy number reduction of our gadB overexpression cassette plasmids (accomplished by replacing the ColE1 origin with the p15A origin) was expected to decrease the overall amount of gadB expression, thereby decreasing the metabolic load imposed on the cells and the mutation rates observed in the gadB gene. However, we also observed several mutations within the gadB gene in the potentially positive cassette plasmids sequenced. The observed mutations are detailed in Table 2.
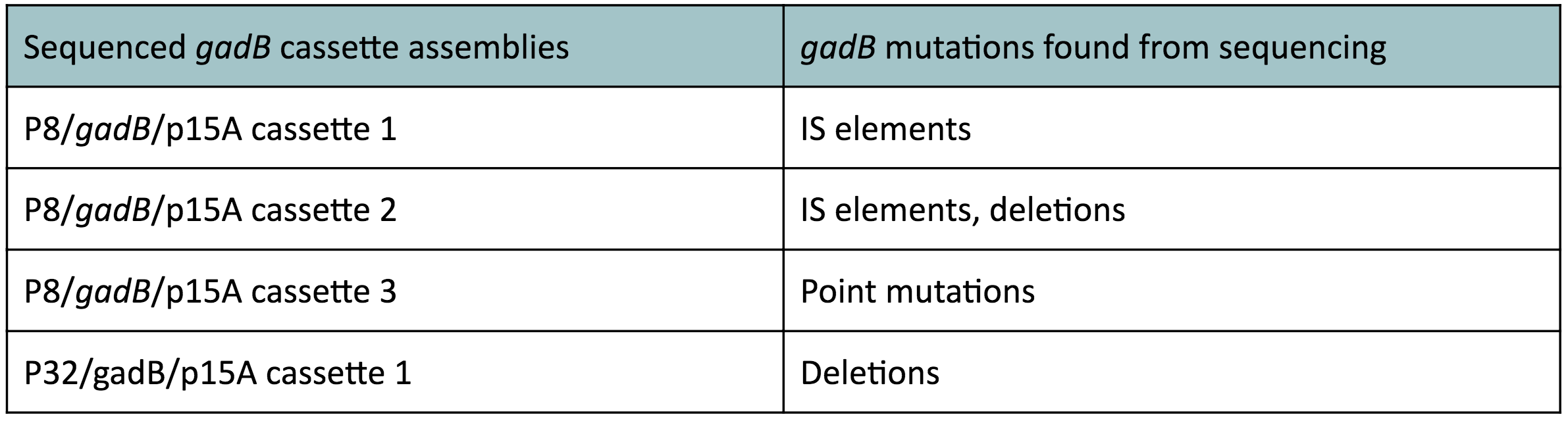
Table 2 gadB mutations in sequenced P8/
gadB/p15A and P32/
gadB/p15A overexpression cassette plasmids.
Given our experimental results and the fact that inducibly expressed genetic devices are more evolutionarily stable than constitutively expressed ones (6), we attempted to inducibly express the gadB gene using the regulatory elements of the lac operon to see if expression and maintenance of a stable gadB gene was possible in E. coli. Our IPTG-inducible gadB expression cassette plasmid was assembled using a lac promoter and operator part plasmid, the gadB gene part plasmid, the M13 terminator part plasmid, connector part plasmids, a CP25 promoter part plasmid, a lacI part plasmid, and the SpecR and p15A origin part plasmid (Fig. 8). Under this assembled regulatory system, in the absence of IPTG (an analog of the allolactose inducer) the LacI repressor will bind to the lac operator region to block transcription of the gadB gene. When present, IPTG will act as an inducer and bind to the LacI repressor to decrease its binding affinity for the lac operator, thereby allowing for gadB expression. This IPTG-inducible system provides us with a mechanism of controlling gadB expression. Positive colonies have been identified and sequence verification is currently underway.
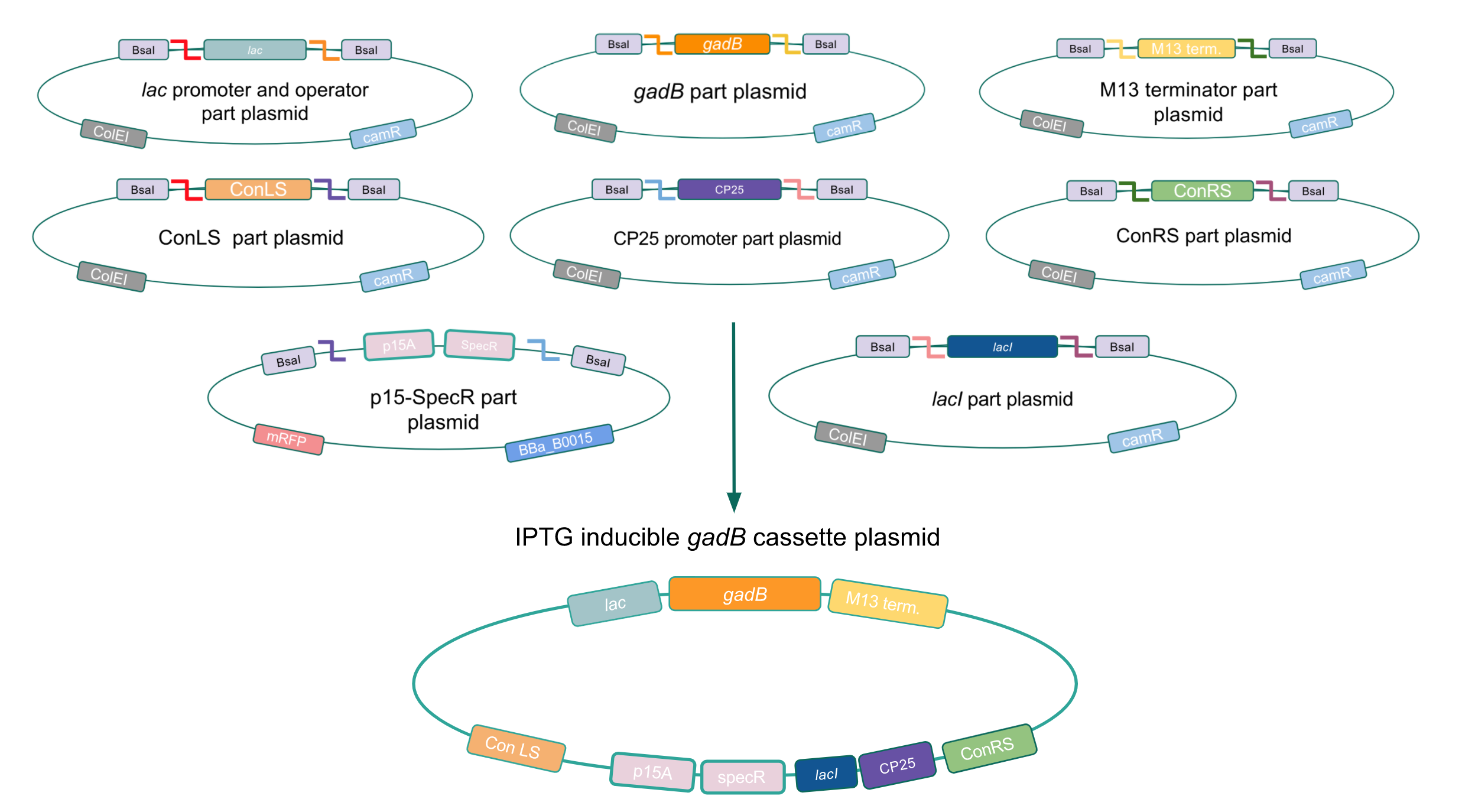
Figure 8. Golden Gate assembly process of IPTG-inducible
gadB expression cassette plasmid.
Golden Gate Compatible Shuttle Vector
We wanted to assemble our final GABA overexpression cassette plasmid using the shuttle vector pMSP3535 as the backbone (Fig. 9). To do this, we first needed to make pMSP3535 Golden Gate compatible (i.e. free of BsaI restriction sites and containing correct overhangs for cassette assembly). We chose to work with pMSP3535 as it contains both a ColE1 origin for replication in E. coli and a pAMb1 origin for replication in Gram-positive bacteria including Lactobacillus species (7), allowing us to easily transplant our cassette plasmid from E. coli to Lactobacillus plantarum once assembled. Additionally, the pMSP3535 vector contains the resistance gene for erythromycin, of which Lactobacillus plantarum is naturally susceptible (8).
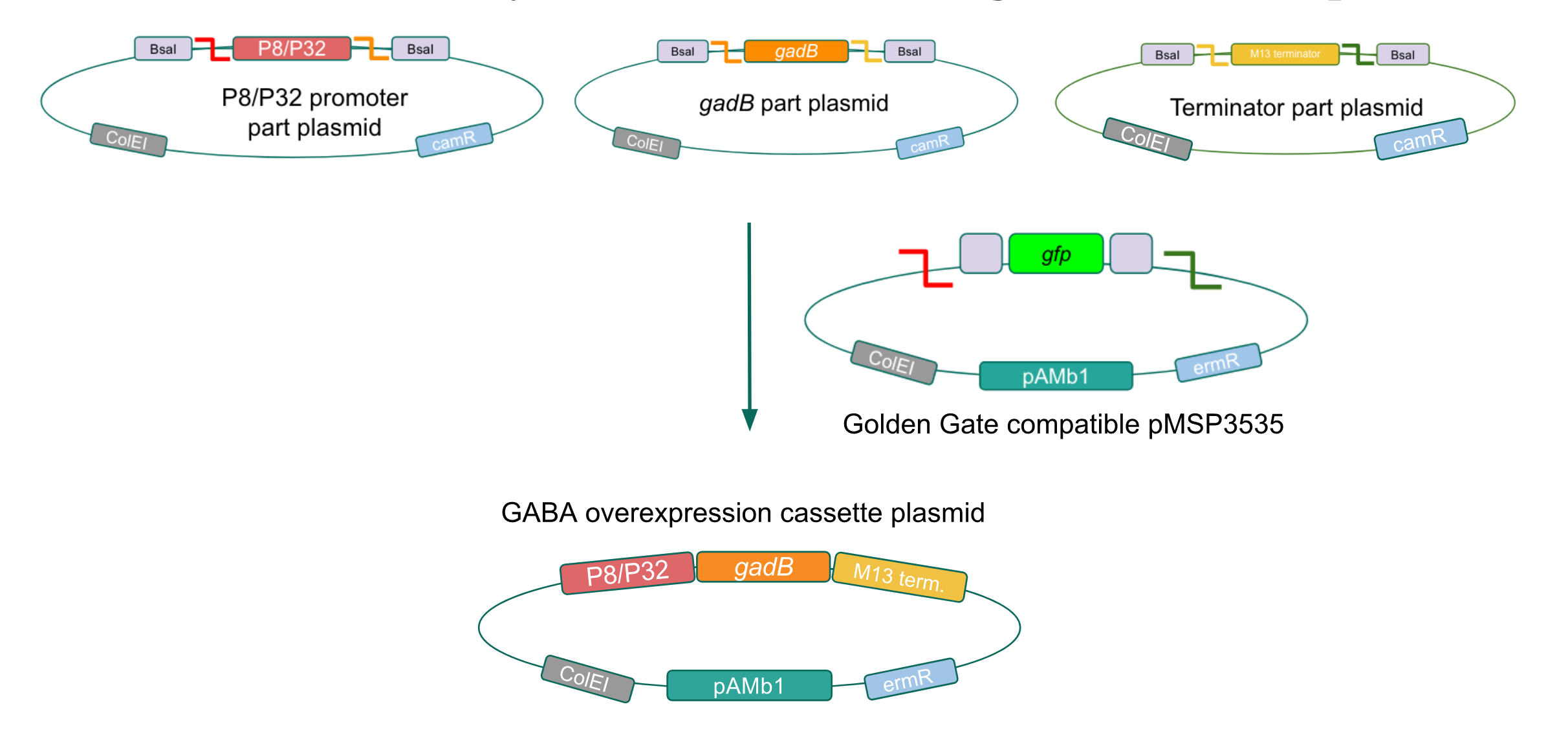
Figure 9. Golden Gate assembly of the GABA final overexpression cassette plasmid with the Golden Gate compatible pMSP3535 vector and the P8/P32 promoter,
gadB gene, and M13 terminator part plasmids.
The process of making the pMSP3535 vector Golden Gate compatible involved two steps: 1) assembling the pMSP3535 backbone (pAMb1 origin and erythromycin resistance gene) with a new ColE1 origin; 2) assembling a gfp dropout part to the assembly of the pMSP3535 backbone and the new ColE1 origin (Fig. 10).
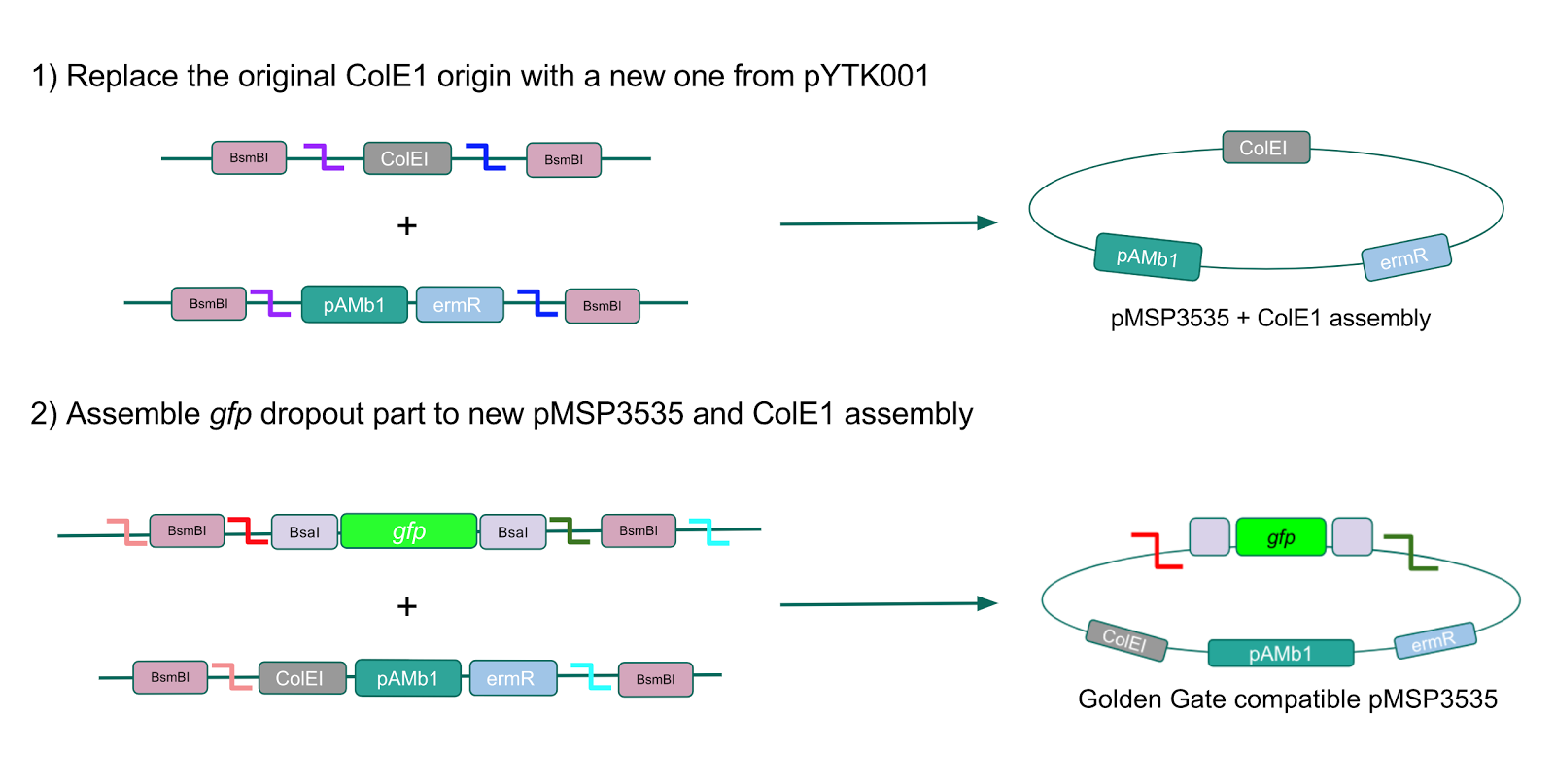
Figure 10. Assembly workflow for creating the Golden Gate compatible pMSP3535 backbone. The first step involves replacing the ColE1 origin in the pMSP3535 backbone with a new ColE1 origin from pYTK001 to create a pMSP3535 + ColE1 assembly. The second step involves combining the pMSP3535 + ColE1 assembly with a
gfp dropout to form the final Golden Gate compatible vector to be used to create our GABA overexpression plasmid.
Since the original pMSP3535 vector contained two illegal BsaI sites within the ColE1 origin, we sought to replace this ColE1 origin with a BsaI-free one isolated from pYTK001. This assembly process involved linearizing and adding BsmBI sites and compatible overhangs to the pMSP3535 backbone and the pYTK001 ColE1 origin via PCR. After the pMSP3535 backbone and ColE1 origin were successfully amplified by PCR (Fig. 11a), they were joined together using BsmBI assembly. Diagnostic PCR was performed on pMSP3535 + ColE1 minipreps from E. coli transformants to screen for positive samples containing both the pMSP3535 backbone and the ColE1 inserts (Fig. 11b). After confirming the presence of the pMSP3535 vector and ColE1 origin, we partially sequence-confirmed the two miniprep samples.
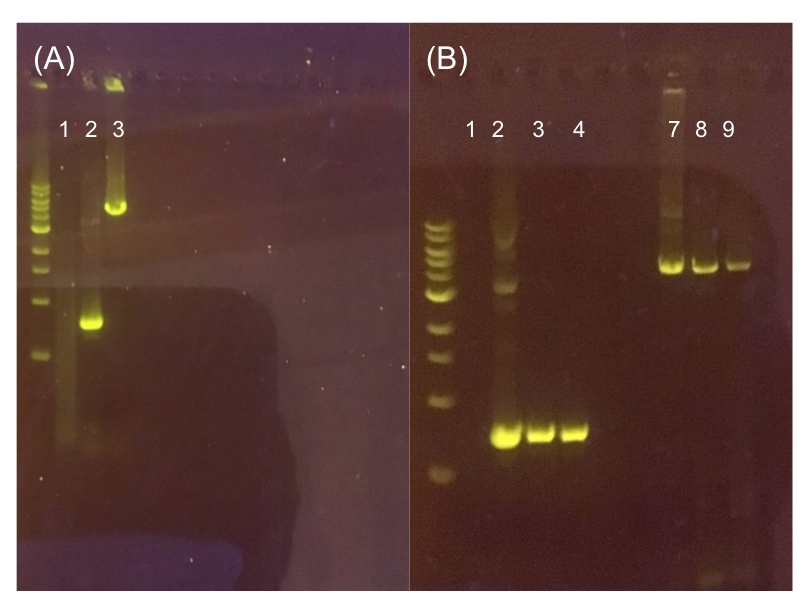
Figure 11. (A) Agarose gel of successful PCR for the pMSP3535 backbone and the ColE1 origin. The ladder used is the 1KB NEB ladder. Lane 1 contains a negative PCR control. Lanes 2 and 3 contain the ColE1 origin (around 800 bp) and pMSP3535 backbone PCR products (around 4.3 kb). (B) Agarose gel of diagnostic PCR of miniprep samples from pMSP3535 + ColE1 transformants. Lane 1 contains a negative PCR control. Lane 2 contains a positive PCR control for the pMSP3535 backbone, while lanes 3 and 4 contain the pMSP3535 PCR from two miniprep samples. Lane 7 contains a positive PCR control for the ColE1 origin, while lanes 8 and 9 contain the ColE1 PCR from the same two miniprep samples. Evidently, the pMSP3535 backbone and ColE1 origin is present in both tested miniprep samples.
To this pMSP3535 + ColE1 assembly, we wanted to add a gfp dropout part containing internal BsaI sites that will generate overhangs compatible with those in the P8/P32 promoter and M13 terminator part plasmids. Additionally, the incorporation of this gfp dropout part will also allow us to visually screen for positive and negative transformants based on their fluorescence. BsmBI sites and compatible overhangs were added to the gfp dropout part by PCR amplifying it from pYTK047. We have been attempting to linearize and add BsmBI sites and overhangs to the positive pMSP3535 + ColE1 assemblies via PCR, with no success. However, results from diagnostic digests suggested that our assemblies may have contained extra, undesired DNA such as IS elements (Fig. 12). Thus, as of right now, we are screening for more positive pMSP3535 + ColE1 transformants. Once we have trouble-shooted this problem, the pMSP3535 + ColE1 and the gfp dropout PCR products will be joined through BsmBI assembly to form the final Golden Gate compatible pMSP3535 vector.
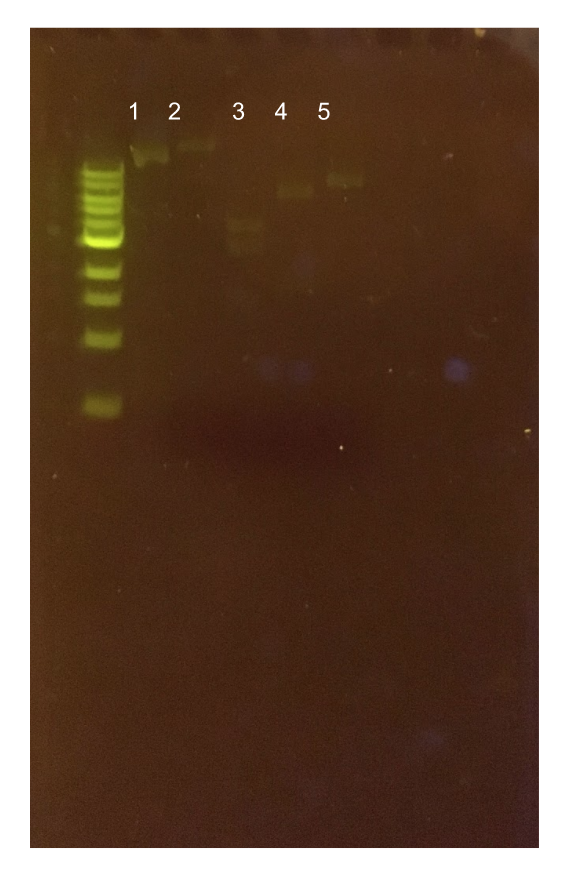
Figure 12. Agarose gel of pMSP3535 + ColE1 assembly diagnostic digests. The ladder used is the 1KB NEB ladder. Lane 1 contains the undigested plasmid assembly. Lane 2 contains the ClaI-digested plasmid with expected 400 bp and 4.6 kb bands. The actual band generated is apparently above 10 kb. Lane 3 contains the XmnI-digested plasmid with expected 200 bp, 1.5 kb, and 3.3 kb bands. The generated band sizes were 2.5 kb and 4 kb. Lane 4 contains the KpnI-digested assembly with an expected 5.1 kb band. The generated band size was 6 kb. Lane 5 contains the Bg1II-digested assembly with an expected 5.1 kb band. The generated band size was 8 kb.
Assessing erythromycin susceptibility of E. coli
Because we are creating our Golden Gate compatible pMSP3535 shuttle vector in E. coli, we wanted to determine the natural susceptibility of E. coli to erythromycin as the minimum concentration to use has not been established clearly in the literature. Thus, we performed an erythromycin minimum inhibitory concentration test in liquid LB media (Fig. 13). After one-day incubation, we observed that E. coli was resistant up to around 150 µg/mL of erythromycin. From this experiment, we have determined that the optimal erythromycin concentration for selecting against E. coli in liquid culture is around 200-250 µg/mL.

Figure 13. Erythromycin minimum inhibitory concentration tests for
E. coli in liquid media. From left to right: 0 µg/mL, 50 µg/mL, 100 µg/mL, 150 µg/mL, 200 µg/mL, 250 µg/mL, 300 µg/mL, 350 µg/mL, and 400 µg/mL.
Back to Top
Lactobacillus plantarum transformation
Lactobacillus plantarum is a gram-positive lactic acid producing bacteria, so it requires a different growth media than we typically use in our lab. In 1954, Briggs agar was developed (9). This media was designed for Lactobacilli, but was not sufficient for many species, including Lactobacillus plantarum , so a different non-selective media for general Lactobacilli was developed in 1960 by Man, Rogosa and Sharpe and named MRS (10). We have exclusively grown our Lactobacillus plantarum on MRS media. Further, we grew Lactobacillus plantarum in a CO2 incubator as referenced in most literature we studied (11-13). The metabolic pathways in the bacteria alter when grown aerobically to produce excess acetate (14) and less lactic acid. Because we intend to utilize this bacteria in a fermentable food, a change in this metabolic pathway would not benefit our ultimate goal.Click here for our MRS media recipe
Once we could successfully grow Lactobacillus plantarum, we wanted to transform it with pMSP3535. In order to do this, we identified and worked with a different protocol than we had ever used in our lab. We attempted several protocols, including Landete 2014 (11) and Speer 2012 (12). However, we found success using a variation of the Welker protocol(13). Welker et al. transformed multiple strains of Lactobacillus casei using varying reagents and yielded different efficiencies between each strain of the species with each variation.
The Texas Tech iGEM team helped us by testing the Speer protocol after we had attempted three procedures and hadn't successfully transformed. They were not able to successfully transform using the Speer protocol which suggests that the procedure was either too simplified for researchers who have never transformed gram-positive bacteria, or was not compatible with Lactobacillus plantarum.Click here for our electroporation protocol
After preparing the necessary solutions, we followed the Welker protocol with some minor differences. We inoculated bacterial stocks with 10 mL of MRS broth in a CO2 incubator, without shaking, overnight. After this, we subcultured the bacteria in 200 mL of prewarmed MRS broth with 0.9M NaCl from an OD600 of 0.1 until it reached an OD600 of 0.6 (~6 hours). After harvesting the cells by centrifugation and rinsing, we resuspended the cells in 4 mL of cold water and eight 0.5 mL aliquots were divided into 1.5 mL microcentrifuge tubes.
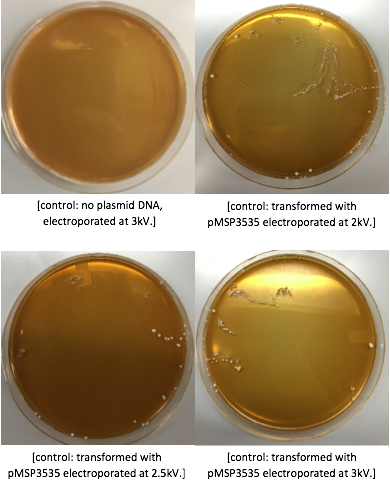
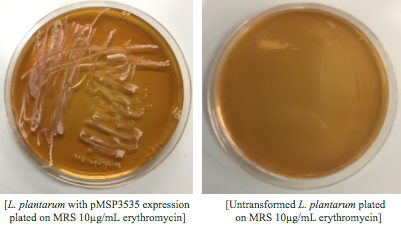
The cells were washed several more times with water and 30% PEG 8000, then stored in a -80°C freezer. These cells should stay viable for up to two years.To enhance the transformation efficiency, 600 μL of prepared cells were suspended 900 μL of cold, sterile, distilled water for 30 min as a pretreatment to electroporation.The cells were pelleted and washed several times with water and 30% PEG 8000. We added 100 ng of plasmid DNA to 100μL of cell suspension as opposed to 200 ng that the protocol recommends; our first plasmid of choice was pMSP3535. This plasmid expresses erythromycin resistance. The cells were electroporated in 2mm cuvettes using the Ec3 settings on the BioRad electroporator which corresponds to 600Ω and 3 kV, differing from the Welker protocol recommendations.
After cells were electroporated, they recovered overnight in the appropriate recovery media. They were plated on MRS agar plates with 10μg/mL erythromycin and left to grow for 2 days.
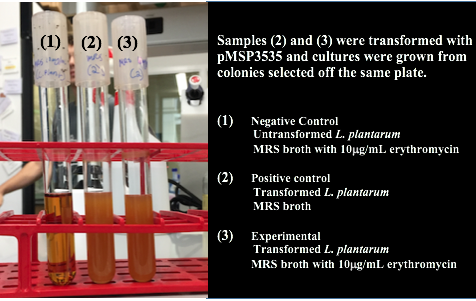
Colonies from each transformation plate were grown up in MRS broth supplemented with 10μg/mL erythromycin. The subsequent day, they were streaked on 10μg/mL erythromycin MRS agar plates to verify resistance.
Genomic and plasmid sequence verification is underway.
We aimed to show a visible transformation of Lactobacillus plantarum in addition to the resistance, so we transformed again using a different plasmid. This time we transformed with pBTK501, a plasmid that codes for a resistance to ampicillin and expresses gfp.
References
- Yunes, R. A et al. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe. 42: 197-204 (2016).
- Zhu, D. et al. Isolation of strong constitutive promoters from Lactococcus lactis subsp. Lactis N8. FEMS Microbiol Lett. 363(16): pii: fnv107 (2015).
- Lee, M. E. et al. A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 4(9): 975-86 (2015).
- Commichau, F. M., et al. Glutamate metabolism in Bacillus subtilis: Gene Expression and Enzyme Activities Evolved To Avoid Futile Cycles and to allow Rapid Responses to Perturbations of the System. AMS. 190(10): 3557-64 (2008).
- Sarkar, P.K. and Nout, M.J.R. (Eds.) 2014. Handbook of Indigenous Foods Involving Alkaline Fermentation. CRC Press/Taylor & Francis Group, p. 629.
- Sleight, S. C. et al. Designing and engineering evolutionary robust genetic circuits. Journal of Biological Engineering. (4):12 (2010).
- Pérez-Arellano, I. et al. Construction of Compatible Wide-Host-Range Shuttle Vectors for Lactic Acid Bacteria and Escherichia coli. Plasmid. 46(2): 106-16 (2001).
- Flórez A. B. et al. Susceptibility of Lactobacillus plantarum Strains to Six Antibiotics and Definition of New Susceptibility–Resistance Cutoff Values. Microbial Drug Resistance. 12(4): 252-56 (2007).
- Cox, C. P., & Briggs, M. (1954). Experiments On Growth Media For Lactobacilli. Journal of Applied Bacteriology, 17(1), 18-26. doi:10.1111/j.1365-2672.1954.tb02019.x
- Man, J. C., Rogosa, M., & Sharpe, M. E. (1960). A Medium For The Cultivation Of Lactobacilli. Journal of Applied Bacteriology, 23(1), 130-135. doi:10.1111/j.1365-2672.1960.tb00188.x
- Speer, M., & Richard, T. (n.d.). Lactobacillus transformation (Speer 2012). Retrieved May 15, 2017, from https://openwetware.org/wiki/Lactobacillus_transformation_(Speer_2012)
- Landete, J. M., Arqués, J. L., Peirotén, Á, Langa, S., & Medina, M. (2014). An improved method for the electrotransformation of lactic acid bacteria: A comparative survey. Journal of Microbiological Methods, 105, 130-133. doi:10.1016/j.mimet.2014.07.022
- Dennis L. Welker, Joanne E. Hughes, James L. Steele, Jeff R. Broadbent; High efficiency electrotransformation of Lactobacillus casei, FEMS Microbiology Letters, Volume 362, Issue 2, 1 January 2015, Pages 1–6, https://doi.org/10.1093/femsle/fnu033
- Murphy, M. G., & Condon, S. (1984). Comparison of aerobic and anaerobic growth of Lactobacillus plantarum in a glucose medium. Archives of Microbiology, 138(1), 49-53.
Back to Top