iGEM AQA_Unesp
results
editing

Figure 1. Electrophoresis gel for the cloned plasmids: pSEUDO_GFP (Ctrl), pSEUDO_174 (174) and pSEUDO_175.
Lactococcus lactis
We also had success at 177, 179 and 180 sequences in E. coli TOP 10 and Lactococcus lactis cremoris as shown in figure 2 and 3.

Figure 2. Electrophoresis gel for the cloned plasmids: pSEUDO_177 and pSEUDO_180.

Figure 3. Gene sequence alignment executed in Serial Cloner, comparing obtained sequence by capillary electrophoresis and planned nucleotide sequence.
In order to detect insulin production, we made an western blot (targeting His6-tag) by selecting the supernatant of a 50 mL penetratin_SCI-57_His6 (177) producer Lactococcus lactis culture. Despite of the “noisy” band, we concluded that this strain is capable of producing and secreting insulin and the use of dry milk might have interfered at the band detection [1]. In the future, more western blotting procedure could elucidate this hypothesis.

Figure 4. Western Blot membrane for the cloned plasmids: pSEUDO_GFP (Ctrl) and pSEUDO_177 (177).
Bacillus subtilis
Bacillus subtilis K07 transformation was also achieved as shown in figure 5.

Figure 5. Western Blot membrane for the cloned plasmids: pSEUDO_GFP (Ctrl) and pSEUDO_177 (177).
To detect our peptide production and secretion, we’ve made a SDS-Page of a 50 mL 182 producer Bacillus subtilis K07 culture supernatant, right after its clarification with a 5 kDa cut-off membrane filter. We can see, through the figure 6, that only pLike-rep transformed strain presented a few insulin production.
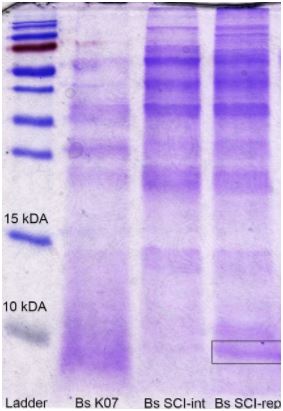
Figure 6. SDS-page gel for cloned plasmids: Bs K07 (control), pBs1C (Bs SCI-int) and pLike-rep (Bs SCI-rep).
characterization
Growth and GFP production
We have characterized bacterial growth and gene reporter production influence of two promoters: srfA (for Bacillus subtilis) and usp45 (for Lactococcus lactis).
Figure 7. Growth curve and fluorescence from Bacillus subtilis K07 cloned with Prsfa_GFP genes.

Figure 8. Growth curve and fluorescence from Lactococcus lactis cremoris cloned with Pusp45_GFP genes.
Through the presented graphics on figures 7 and 8, we could assume that both srfa and usp45 promoters do not interfere in the bacterial growth and GFP production follows the same cellular growth behavior, presenting the most product accumulation in the stationary phase.
sRNA
After cloning the 175 (Pgal_sRNA_termrnnb (Ec)) sRNA sequence, we tested its behavior and efficiency when associated at different carbon sources, such as glucose, fructose, mannitol (a sugar alcohol) and galactose.

Figure 9. Relative fluorescence (Fluorescence/OD) of sRNA (175) producers and non-producers (Ctrl) in different carbon sources.
After analysing the graphic in figure 9, we can admit that glucose and fructose presence “turned ON” the gene reporter expression, showing similar relative fluorescence values to the control. Meanwhile, when exposed to other carbon sources, such as mannitol and galactose, the gene reporter expression was “turned OFF”, presenting then lower relative fluorescence values, when compared with the control. This result shows that the sRNA design was successful, but further characterization should be executed for better understanding.
References
[1] MAHMOOD, Tahrin; YANG, Ping-Chang. Western blot: technique, theory, and troubleshooting. North American journal of medical sciences, v. 4, n. 9, p. 429, 2012.


