| (130 intermediate revisions by 5 users not shown) | |||
| Line 3: | Line 3: | ||
<div id="pagebanner" style="background-image: url(https://static.igem.org/mediawiki/2017/3/3e/T--TU_Dresden--planet--communication.png);"> | <div id="pagebanner" style="background-image: url(https://static.igem.org/mediawiki/2017/3/3e/T--TU_Dresden--planet--communication.png);"> | ||
| − | <div | + | <div> |
| − | + | <div id="bannerquote">Contacting Extra-Peptidosomal Life</div> | |
| − | + | <div id="projecttitle">Communication</div> | |
| − | + | ||
| + | <div class="contentbox at-a-glance"> | ||
| + | <h1 class="box-heading">At a Glance</h1> | ||
| + | <figure> | ||
| + | <figure class="makeresponsive floatright" style="width: 33%;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/3/3b/T--TU_Dresden--P_Communication_at_a_glance.png" class="zoom"></figure> | ||
| + | <h4>Motivation:</h4> | ||
| + | <p>Demonstrate that two different strains of encapsulated bacteria could exchange soluble compounds across the Peptidosome barrier.</p> | ||
| + | <p></p> | ||
| + | <h4>Approach:</h4> | ||
| + | <p>Develop a sender-receiver strain pair in <i>Bacillus subtilis</i> that communicates via peptide-dependent quorum sensing.</p> | ||
| + | <p></p> | ||
| + | <h4>Achievements:</h4> | ||
| + | <p>(I) A functional <a href="#sestrest" class="hashlink">sender-receiver</a> pair was established, based on the ComX pheromone and the ComP-ComA-P<sub><i>srfA</i></sub> signaling system. (II) The sender could activate the receiver in <a href="#coculture" class="hashlink">co-culturing</a> experiments. (III) The same behavior was observed in <a href="#peptidosomes" class="hashlink">Peptidosome</a> cultivations. (IV) 6 novel basic <a href="#BioBricks" class="hashlink">BioBrick parts</a> were generated and fully evaluated for functionality.</p> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <main> | ||
<div class="contentbox"> | <div class="contentbox"> | ||
| − | <h1 class="box-heading"> | + | <h1 id="communication" class="box-heading">Short Description</h1> |
<p> | <p> | ||
| − | By using Peptidosomes we introduce a new powerful platform for co-culturing. This technique physically separates bacterial populations without limiting their ability to communicate with each other via | + | By using Peptidosomes we introduce a new powerful platform for co-culturing. This technique physically separates bacterial populations without limiting their ability to communicate with each other via signaling molecules. This part of EncaBcillus is focused on proving the concept of communication between encapsulated bacteria by making use of the native regulatory system for competence development in <i>Bacillus subtilis</i> which is based on quorum sensing. |
</p> | </p> | ||
</div> | </div> | ||
<div class="contentbox"> | <div class="contentbox"> | ||
| − | <h1 class="box-heading">Background</h1> | + | <h1 id="background" class="box-heading">Background</h1> |
<p> | <p> | ||
| Line 40: | Line 58: | ||
<p> | <p> | ||
| − | The well-studied regulatory system for competence development in <i>B. subtilis</i> provided a genetic set-up based on quorum sensing that we used as a proof of principle <a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/12576575">[4]</a> | + | The well-studied regulatory system for competence development in <i>B. subtilis</i> provided a genetic set-up based on quorum sensing that we used as a proof of principle (Figure 1). <a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/12576575">[4]</a> |
| − | <i>B. subtilis</i> constantly secretes the ComX pheromone, a 9- to 10-amino acid oligopeptide, as a | + | <i>B. subtilis</i> constantly secretes the ComX pheromone, a 9- to 10-amino acid oligopeptide, as a signaling molecule <b>(a)</b>. By rising cell-density, the ComX-concentration in the surrounding medium increases until it reaches a threshold and activates ComP, a membrane-spanning protein kinase <b>(b)</b>. |
The kinase reacts to the accumulation of ComX by phosphorylating the response regulator ComA <b>(c)</b> which then works as a transcription factor by binding to several promoters and enhancing their activity <b>(d)</b> | The kinase reacts to the accumulation of ComX by phosphorylating the response regulator ComA <b>(c)</b> which then works as a transcription factor by binding to several promoters and enhancing their activity <b>(d)</b> | ||
<a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/26582911">[5</a>,<a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/18585392">6]</a>.</p> | <a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/26582911">[5</a>,<a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/18585392">6]</a>.</p> | ||
| Line 53: | Line 71: | ||
<h3>Communication between encapsulated bacteria and bacteria in the surroundings </h3> | <h3>Communication between encapsulated bacteria and bacteria in the surroundings </h3> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | <figure class="makeresponsive" style=" | + | <figure> |
| + | <figure class="makeresponsive floatright" style="width:60%;"> | ||
<img src="https://static.igem.org/mediawiki/2017/6/65/T--TU_Dresden--P_Communication_Background.png" | <img src="https://static.igem.org/mediawiki/2017/6/65/T--TU_Dresden--P_Communication_Background.png" | ||
alt="Figure 1 Communication" class="zoom"> | alt="Figure 1 Communication" class="zoom"> | ||
| − | <figcaption><b>Figure 2: Illustration of co-cultured sender (ReSt) and receiver strain (SeSt) using Peptidosomes.</b> The SeSt in the surrounding medium secretes the ComX quorum sensing pheromone that diffuses through the membrane into the Peptidosome and stimulates the encapsulated, comX-deficient ReSt. As a result the ReSt produces luminescence driven by | + | <figcaption><b>Figure 2: Illustration of co-cultured sender (ReSt) and receiver strain (SeSt) using Peptidosomes.</b> The SeSt in the surrounding medium secretes the ComX quorum sensing pheromone that diffuses through the membrane into the Peptidosome and stimulates the encapsulated, comX-deficient ReSt. As a result, the ReSt produces luminescence driven by a ComX-dependant promoter upstream the lux operon.</figcaption> |
| + | </figure> | ||
| + | <p> | ||
| + | Based on this genetic background, we wanted to set up a simple experiment that shows communication between two <i>B. subtilis</i> strains. The basic idea is comparable to a radio broadcast: you need a transmitter that sends out the radio waves and a receiver that receives the waves and converts them into an acoustical signal. Transferring this idea over to our project means that we need a sender strain (SeSt) that secretes the ComX pheromone, and a receiver strain (ReSt) that reacts to the ComX-stimulation with an easy-detectable output (Figure 2).</p> | ||
| + | <p> | ||
| + | In our setup, we fused target promoters of the competence machinery of <i>B. subtilis</i> to the <i>lux</i> operon and measured luminescence output. Additionally, the ReSt needed to be <i>comX</i>-deficient to prevent autoinduction. We encapsulated the ReSt within Peptidosomes and incubated it together with the SeSt (either wild type or a ComX-overproducing strain) in the surrounding medium. Thus, we expected ComX that is produced outside the Peptidosme to diffuse through the membrane and stimulate the ReSt. As a result, we should obtain a glowing Peptidosome mediated by the <i>lux</i> operon, which is only present in the ReSt.</p> | ||
</figure> | </figure> | ||
</div> | </div> | ||
| Line 68: | Line 88: | ||
| − | <div class="contentbox"> | + | <div id="design" class="contentbox"> |
<h1 class="box-heading">Design</h1> | <h1 class="box-heading">Design</h1> | ||
| − | <p>After extensive research, an iGEM project starts with designing genetic constructs and planning experiments. It takes some time until the strains are finished and the first assays are running. Anyway, it is no secret that the most important results come in the last few weeks before the Giant Jamboree. Figure 3 gives an overview of this subproject in | + | <p>After extensive research, an iGEM project starts with designing genetic constructs and planning experiments. It takes some time until the strains are finished and the first assays are running. Anyway, it is no secret that the most important results come in the last few weeks before the Giant Jamboree. Figure 3 gives an overview of this subproject in chronological order.</p> |
| − | <figure class="makeresponsive" style="padding-left: | + | <figure class="makeresponsive" style="padding-left: 25%; padding-right: 25%;"> |
<img src="https://static.igem.org/mediawiki/2017/7/73/T--TU_Dresden--P_Communication_Figure3.jpeg" | <img src="https://static.igem.org/mediawiki/2017/7/73/T--TU_Dresden--P_Communication_Figure3.jpeg" | ||
alt="Figure 3 Communication" class="zoom"> | alt="Figure 3 Communication" class="zoom"> | ||
| Line 87: | Line 107: | ||
<h4>Sender strain (SeSt)</h4> | <h4>Sender strain (SeSt)</h4> | ||
| − | + | <figure> | |
| + | <figure class="makeresponsive floatleft" style="width:40%;"> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/7/77/T--TU_Dresden--P_Communication_Pxyl_comX.png" | ||
| + | alt="Figure 4" class="zoom"> | ||
| + | <figcaption><b>Figure 4: Inducible copy of <i><b>B. subtilis</b></i> ComX pheromone.</b></figurecaption> | ||
| + | </figure> | ||
<p>As SeSt, we used <i>B. subtilis</i> W168 (WT) with an additional inducible copy of <i>comX</i> (Table 1) in order to increase the concentration of ComX in the medium independently from cell density. Overproduction of ComX was achieved due to a xylose-inducible promoter (P<sub><i>xylA</i></sub>, Figure 4). For that reason, we used the pBS2EP<sub><i>xylA</i></sub>, an integrative plasmid of the <i>Bacillus</i> BioBrick Box 2.0 (Popp <i>et al.</i> 2017, accepted). </p> | <p>As SeSt, we used <i>B. subtilis</i> W168 (WT) with an additional inducible copy of <i>comX</i> (Table 1) in order to increase the concentration of ComX in the medium independently from cell density. Overproduction of ComX was achieved due to a xylose-inducible promoter (P<sub><i>xylA</i></sub>, Figure 4). For that reason, we used the pBS2EP<sub><i>xylA</i></sub>, an integrative plasmid of the <i>Bacillus</i> BioBrick Box 2.0 (Popp <i>et al.</i> 2017, accepted). </p> | ||
| − | + | </figure> | |
</div> | </div> | ||
| Line 96: | Line 121: | ||
<h4>Receiver strain (ReSt)</h4> | <h4>Receiver strain (ReSt)</h4> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<figure> | <figure> | ||
| − | + | <figure class="makeresponsive floatleft" style="width:40%;"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<img src="https://static.igem.org/mediawiki/2017/f/f7/T--TU_Dresden--communication-py_luxpng.png" | <img src="https://static.igem.org/mediawiki/2017/f/f7/T--TU_Dresden--communication-py_luxpng.png" | ||
alt="Figure 5" class="zoom"> | alt="Figure 5" class="zoom"> | ||
| − | <figcaption><b>Figure 5: Target promoters of the competence machinery of <i><b>B. subtilis</b></i> (P<sub><i><b>X</b></i></sub> = P<sub><i><b>srfA</b></i></sub> or P<sub><i><b>rapA</b></i></sub> or P<sub><i><b>rapF</b></i></sub> or P<sub><i><b>comG</b></i></sub> or P<sub><i><b>comK</b></i></sub> mut) fused to the <i><b>lux</b></i> operon.</b> | + | <figcaption><b>Figure 5: Target promoters of the competence machinery of <i><b>B. subtilis</b></i> (P<sub><i><b>X</b></i></sub> = P<sub><i><b>srfA</b></i></sub> or P<sub><i><b>rapA</b></i></sub> or P<sub><i><b>rapF</b></i></sub> or P<sub><i><b>comG</b></i></sub> or P<sub><i><b>comK</b></i></sub> mut) fused to the <i><b>lux</b></i> operon.</b></figurecaption> |
| + | </figure> | ||
| + | <p>To create a strain suitable as ReSt we screened various promoters that are part of the quorum response to ComX. We have chosen three promoters, P<sub><i>srfA</i></sub>, P<sub><i>rapA</i></sub> and P<sub><i>rapF</i></sub>, that are controlled by the response regulator ComA as well as the the ComK-dependent promoter P<sub><i>comG</i></sub> and the mutated promoter of the competence transcription factor ComK (P<sub><i>comK</i></sub> mut) itself (Table 1). | ||
| + | All promoters were cloned into the pBS3C<i>lux</i>, an integrative plasmid of the <i>Bacillus</i> BioBrick Box containing the <i>luxABCDE</i> reporter to measure promoter activities (Figure 5) <a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/24295448">[9]</a>.</p> | ||
</figure> | </figure> | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
<style> | <style> | ||
.jonathanstables{ | .jonathanstables{ | ||
| Line 133: | Line 147: | ||
} | } | ||
</style> | </style> | ||
| − | <figure class="jonathanstables"> | + | <figure class="jonathanstables" id="BioBricks"> |
| − | <figcaption><b>Table1: BioBricks created for the subproject "Communication".</b></figcaption> | + | <figcaption><b>Table1: Basic BioBricks created for the subproject "Communication".</b></figcaption> |
<table class="makeresponsive"> | <table class="makeresponsive"> | ||
<colgroup> | <colgroup> | ||
| Line 160: | Line 174: | ||
<tr> | <tr> | ||
<td>P<i><sub>comK</sub></i> mut</td><td> | <td>P<i><sub>comK</sub></i> mut</td><td> | ||
| − | <a target="_blank" href="http://parts.igem.org/Part: | + | <a target="_blank" href="http://parts.igem.org/Part:BBa_K2273013">BBa_K2273013</a></td><td>In order to remove a SpeI restriction site we had to exchange one nucleotide (nt 32, A to T).</td></tr> |
</table> | </table> | ||
</figure> | </figure> | ||
| − | <p>All parts have been amplified via PCR from gDNA of <i>Bacillus subtilis </i> W168, verified by sequencing and cloned into the pSB1C3 for storage and submission to the iGEM registry. Cloning was done according to <a href="https://2017.igem.org/Team:TU_Dresden/Experiments">standard protocols</a>. Plasmids have been multiplied with <i>Escherichia coli</i> | + | <p>All parts have been amplified via PCR from gDNA of <i>Bacillus subtilis </i> W168, verified by sequencing and cloned into the pSB1C3 for storage and submission to the iGEM registry. Cloning was done according to <a href="https://2017.igem.org/Team:TU_Dresden/Experiments">standard protocols</a>. Plasmids have been multiplied with <i>Escherichia coli</i> DH10β. All assays have been conducted with <i>B. subtilis</i> W168 carrying one or more of the earlier-described constructs.</p> |
<hr> | <hr> | ||
<h3>Experimental design</h3> | <h3>Experimental design</h3> | ||
| − | |||
<p> | <p> | ||
| − | In order to investigate quorum sensing-dependent effects we determined the promoter activities of P<sub><i>srfA</i></sub>, P<sub><i>rapA</i></sub>, P<sub><i>rapF</i></sub>, P< | + | In order to investigate quorum sensing-dependent effects we determined the promoter activities of P<sub><i>srfA</i></sub>, P<sub><i>rapA</i></sub>, P<sub><i>rapF</i></sub>, P<sub><i>comG</i></sub> and P<i><sub>comK</sub></i> mut in WT and in <i>comX</i>-deficient strains by monitoring the luminescence. In the knockout-strains <i>comX</i> has been replaced with either an erythromycin or a kanamycin resistance cassette (<i>comX::ery</i> or <i>comX::kanR</i>, for simplicity from now on referred to as Δ<i>comX</i>). </p> |
| + | <p>Furthermore, we examined the influence of different media on promoter activity using Luria-Bertani (LB) broth as full medium as well as two minimal media: MCSE and MNGE medium. The latter is commonly used for the <a href="https://2017.igem.org/Team:TU_Dresden/Experiments">transformation</a> of <i>B. subtilis</i>. All plate reader assays (PR-Assays) have been performed with the CLARIOstar® microplate reader from BMG Labtech. | ||
| + | </p> | ||
| + | <p>For co-cultivation of ReSt and SeSt we used either ThinCerts™ - TC inserts (6 well, 0.4 µm pores, transparent) by Greiner or Peptidosomes. Peptidosomes have been prepared according to <a href="https://2017.igem.org/Team:TU_Dresden/Project/Peptidosomes#creation">standard procedures</a>.</p> | ||
| − | + | <div style="display: flex; align-items: flex-start; flex-wrap: wrap;"> | |
| − | + | <div class="makeresponsive" style="width: 40%;"> | |
| − | <div style=" | + | |
| − | < | + | <p>For the co-culture assays with SeSt and ReSt using ThinCerts™ (Figure 6), separate day cultures were inoculated (1:1000) from overnight cultures and incubated at 37°C and 220 rpm until they reached an optical density at 600 nm (OD<sub>600</sub>) of 0.2 – 0.4. From this day cultures, fresh MNGE medium was inoculated to an OD<sub>600</sub> of 0.05 and distributed to the well plate according to the pipette scheme (Table 2) in a final volume of 2 mL for each insert and well respectively. After one hour of incubation (37°C, 200 rpm) SeSt were induced with 1% xylose (if necessary). Luminescence was detected directly after induction (t0) and once every following hour (t1 – t5) using a chemiluminescence imaging system.</p> |
| − | + | ||
| − | < | + | |
| − | </ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| − | < | + | <div class="makeresponsive" style="width: 60%;"> |
| − | < | + | <figure class="makeresponsive floatright" style="width: 100%;"> |
| − | + | <img src="https://static.igem.org/mediawiki/2017/7/78/T--TU_Dresden--P_Communication_Figure4.jpeg" alt="Figure 6 Communication" class="zoom"/> | |
| − | < | + | <figcaption><b>Figure 6:</b> Schematic view of a well plate with ThinCerts™. These inserts can be used to co-culture distinct population and to study possible interactions. The membrane provides a physical barrier for cells whereas molecules are still able to diffuse. (Modified from: <a target="_blank" href="https://www.ncbi.nlm.nih.gov/pubmed/24829281">Goers <i>et al.</i></a>)</figurecaption> </figure> |
| + | <figure class="makeresponsive floatright" style="width: 100%"> | ||
| + | <figcaption><b>Table 2: Pipette scheme for co-culture assay using ThinCerts™.</b> The assay was performed in a 6-well plate. For co-culture of sender strain (SeSt) and receiver strain (ReSt), we used cell culture inserts (A, B, C). Sender Strains were induced with 1% xylose (+xyl) if necessary. For the negative controls (D, E, F) inserts have been omitted.</figurecaption> | ||
| + | <img src="https://static.igem.org/mediawiki/2017/e/e5/T--TU_Dresden--P_Communication_Tabelle_3_Pipettierschema.png" alt="Figure 6 Communication" class="zoom"/> | ||
| + | </div> | ||
| + | </div> | ||
<p> | <p> | ||
Co-culture assays using Peptidosomes were performed as well scan experiments according to <a href="https://2017.igem.org/Team:TU_Dresden/Experiments">"Plate reader well scan Peptidosome"</a> protocol, with the following changes:</p> | Co-culture assays using Peptidosomes were performed as well scan experiments according to <a href="https://2017.igem.org/Team:TU_Dresden/Experiments">"Plate reader well scan Peptidosome"</a> protocol, with the following changes:</p> | ||
| Line 200: | Line 214: | ||
<div class="contentbox"> | <div class="contentbox"> | ||
| − | <h1 class="box-heading">Results</h1> | + | <h1 id="screening" class="box-heading">Results</h1> |
<h3>Promoter activity screening</h3> | <h3>Promoter activity screening</h3> | ||
<p> | <p> | ||
| − | Although quorum sensing induces various changes in gene expression, the engineering of the ReSt initially required an extensive screening of several promoters regarding their ComX-dependent activity (Figure | + | Although quorum sensing induces various changes in gene expression, the engineering of the ReSt initially required an extensive screening of several promoters regarding their ComX-dependent activity (Figure 7).</p> |
| − | < | + | <figure> |
| − | + | <figure class="makeresponsive floatright" style="width:70%;"> | |
| − | <figure class="makeresponsive" > | + | |
<img src="https://static.igem.org/mediawiki/2017/6/6b/T--TU_Dresden--P_Communication_Promoterscreening.png" | <img src="https://static.igem.org/mediawiki/2017/6/6b/T--TU_Dresden--P_Communication_Promoterscreening.png" | ||
alt="Figure 7 Promoters" class="zoom"> | alt="Figure 7 Promoters" class="zoom"> | ||
<figcaption><b>Figure 7: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub>, P<sub><i><b>rapA</b></i></sub>, P<i><sub><b>rapF</b></sub></i>, P<i><sub><b>comG</b></i></sub> and P<i><sub><b>comK</b></sub></i> mut (B) in MNGE medium.</b> Wild type (WT, pink) and <i>comX</i>-knockout strains (Δ<i>comX</i>, dark blue) contain a promoter-<i>luxABCDE</i> fusion. </figcaption> | <figcaption><b>Figure 7: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub>, P<sub><i><b>rapA</b></i></sub>, P<i><sub><b>rapF</b></sub></i>, P<i><sub><b>comG</b></i></sub> and P<i><sub><b>comK</b></sub></i> mut (B) in MNGE medium.</b> Wild type (WT, pink) and <i>comX</i>-knockout strains (Δ<i>comX</i>, dark blue) contain a promoter-<i>luxABCDE</i> fusion. </figcaption> | ||
</figure> | </figure> | ||
| − | + | <p>Since the ComX pheromone itself does not act as a transcription factor we examined promoters that are either under the control of the response regulator ComA (P<sub><i>srfA</i></sub>, P<sub><i>rapA</i></sub> & P<sub><i>rapF</i></sub>) or regulated by the competence transcription factor ComK (P<i><sub>comK</sub></i> and P<i><sub>comG</sub></i>). Surprisingly P<sub><i>rapA</i></sub> and P<sub><i>rapF</i></sub> seem to be constitutive as well as P<i><sub>comK</sub></i> mut that is regulated by ComK itself. The unexpected behaviour of P<i><sub>comK</sub></i> mut could be a consequence of the single nucleotide exchange that was necessary to remove a restriction site since small changes of a promoter sequence can influence the binding of regulatory elements. After an initial decrease of activity P<i><sub>comG</sub></i> exhibits a weak increase of activity during stationary phase. However, for our purpose the most promising promoter is P<sub><i>srfA</i></sub>. Although it possesses a high basal activity in the wild type (WT) it reveals a tenfold increase of expression during the transition to stationary phase. In the associated <i>comX</i>-knockout strain (Δ<i>comX</i>) the promoter activity only shows basal activity. This clearly demonstrates the ComX-dependent expression of the <i>srfA</i> operon. | |
| + | </p> | ||
| + | </figure> | ||
<hr> | <hr> | ||
| − | <h3>Influence of different media on the activity of P<sub><i><b>srfA</b></i></sub></h3> | + | <h3>Influence of different media on the activity of P<sub><i><b>srfA</b></i></sub></h3> |
| − | < | + | <figure> |
| − | + | <figure class="makeresponsive floatright" style="width:65%" > | |
| − | <figure class="makeresponsive" style=" | + | |
<img src="https://static.igem.org/mediawiki/2017/e/e4/T--TU_Dresden--P_Communication_srfa.png" | <img src="https://static.igem.org/mediawiki/2017/e/e4/T--TU_Dresden--P_Communication_srfa.png" | ||
alt="Figure 8 Promoters" class="zoom"> | alt="Figure 8 Promoters" class="zoom"> | ||
| − | <figcaption><b>Figure 8: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub> (B) in different media.</b> Wild type (WT, pink) and <i>comX</i>-knockout strains (<i> | + | <figcaption><b>Figure 8: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub> (B) in different media.</b> Wild type (WT, pink) and <i>comX</i>-knockout strains (Δ<i>comX</i>, dark blue) contain a P<sub><i>srfA</i></sub> <i>luxABCDE</i> fusion. </figcaption> |
| + | </figure> | ||
| + | <p>We continued by examining the influence of different media on the activity of P<sub><i>srfA</i></sub>. This step is important because competence development is not only dependent on quorum sensing but also on nutritional conditions. As expected MNGE, the transformation medium of <i>B. subtilis</i>, turned out to be the most convenient medium for our purposes (Figure 8). In rich medium (LB) we observed the lowest promoter activity for the knockout strain as well as the lowest increase of activity in wild type. However, in both minimal media (MCSE and MNGE), we could observe a higher increase of luminescence, although there is a temporary decline of activity after 3 hours of incubation in MCSE-medium. Therefore, we decided to continue using MNGE-medium for all following experiments.</p> | ||
</figure> | </figure> | ||
| − | |||
<hr> | <hr> | ||
| − | <h3>Overproduction of ComX</h3> | + | <h3 id="sestrest">Overproduction of ComX</h3> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | <figure class="makeresponsive | + | <figure> |
| − | <img src="https://static.igem.org/mediawiki/2017/ | + | <figure class="makeresponsive floatright" style="width:50%"> |
| + | <img src="https://static.igem.org/mediawiki/2017/4/44/T--TU_Dresden--P_Communication_inductionassay.png" | ||
alt="Figure 9 Promoters" class="zoom"> | alt="Figure 9 Promoters" class="zoom"> | ||
| − | <figcaption><b>Figure 9: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub> (B) in dependence | + | <figcaption><b>Figure 9: Growth (A) and promoter activity of P<sub><i><b>srfA</b></i></sub> (B) in dependence on xylose-induced ComX production.</b> Wild type (WT, pink and light blue) and <i>comX</i>-knockout strain (<i>ΔcomX</i>, black and dark blue) contain a P<sub><i>srfA</i></sub> <i>luxABCDE</i> fusion. After 1 hour of incubation (dashed line) the cultures were induced with 1% xylose (light blue and black) or with distilled water (dH2O, pink and dark blue). |
</figcaption> | </figcaption> | ||
| + | </figure> | ||
| + | <p>After screening of promoter activity and media-dependence of P<sub><i>srfA</i></sub> we examined the influence of ComX-overproduction via induction with xylose (Figure 9). In strains with an additional copy of comX (WT), an induction with xylose (+ xylose) did not lead to higher luminescence but to an earlier increase in comparison to the non-induced WT (+ dH2O). In knockout strains (∆<i>comX</i>), we could compensate the <i>comX</i>-deficiency via an induction of ComX-production. Surprisingly in those cultures the luminescence increased even earlier as in WT with an additional induction of the ComX-production. The increase of luminescence in the comX-deficient strain without xylose-induction (∆<i>comX</i> + dH2O) served as negative control. In conclusion, we could demonstrate an impact of ComX-overproduction.</p> | ||
</figure> | </figure> | ||
<hr> | <hr> | ||
| − | <h3>Co-culture of SeSt and ReSt using ThinCerts™</h3> | + | <h3 id="coculture">Co-culture of SeSt and ReSt using ThinCerts™</h3> |
| − | < | + | <figure> |
| − | + | <figure class="makeresponsive floatright" style="width: 60%;" > | |
| − | <figure class="makeresponsive" style=" | + | |
<img src="https://static.igem.org/mediawiki/2017/f/fb/T--TU_Dresden--P_Communication_ThinSert.png" | <img src="https://static.igem.org/mediawiki/2017/f/fb/T--TU_Dresden--P_Communication_ThinSert.png" | ||
alt="Figure 10 Promoters" class="zoom"> | alt="Figure 10 Promoters" class="zoom"> | ||
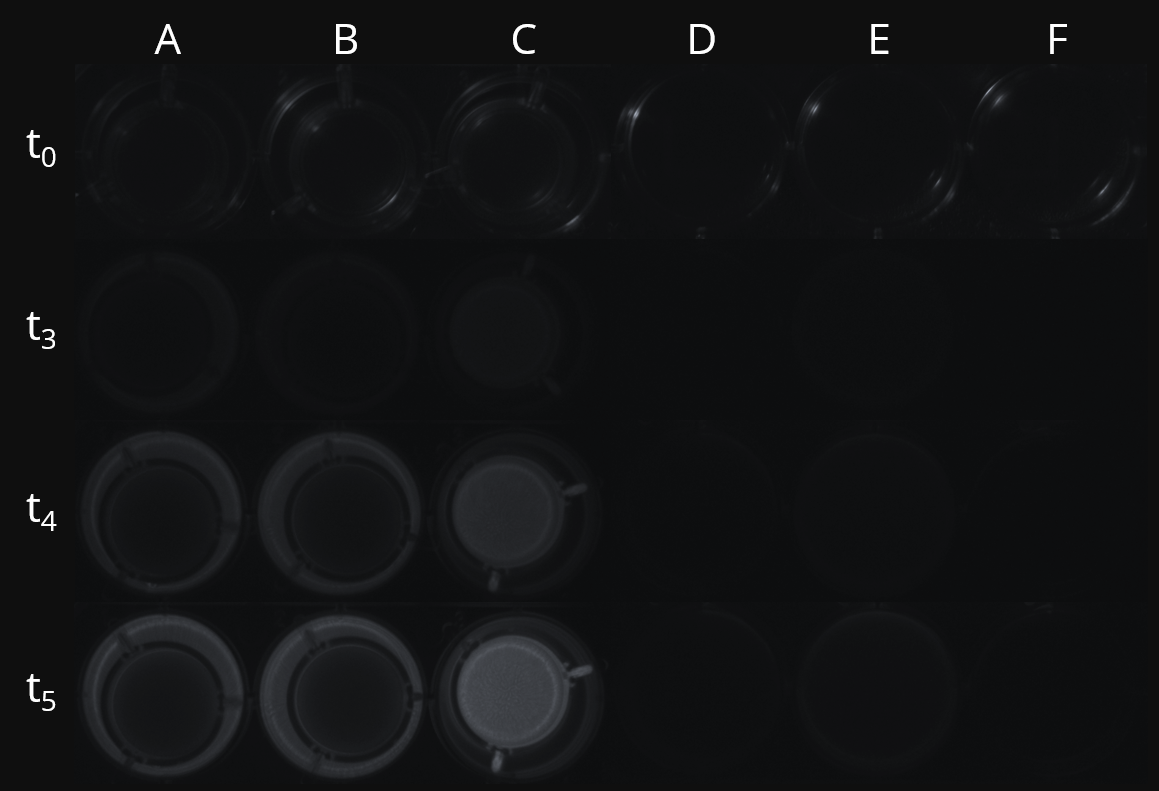
| − | <figcaption><b>Figure 10: Co-culture of sender strain (SeSt) and receiver strain (ReSt) using ThinCert™ cell culture inserts.</b> Luminescence of the ReSt (well) co-cultured with the SeSt (insert) induced with xylose <b>(A)</b>, luminescence of ReSt (well) co-cultured with the SeSt (insert) without xylose-induction <b>(B)</b> and luminescence of ReSt (insert) co-cultured with the SeSt (well) induced with xylose <b>(C)</b> was documented by a chemoluminescence imaging system. Cultures containing only SeSt <b>(D)</b> | + | <figcaption><b>Figure 10: Co-culture of sender strain (SeSt) and receiver strain (ReSt) using ThinCert™ cell culture inserts.</b> Luminescence of the ReSt (well) co-cultured with the SeSt (insert) induced with xylose <b>(A)</b>, luminescence of ReSt (well) co-cultured with the SeSt (insert) without xylose-induction <b>(B)</b> and luminescence of ReSt (insert) co-cultured with the SeSt (well) induced with xylose <b>(C)</b> was documented by a chemoluminescence imaging system. Cultures containing only SeSt <b>(D)</b>, ReSt <b>(E)</b> or MNGE <b>(F)</b> served as negative controls.</figcaption> |
| − | </ | + | </figure> |
| + | <p>Via plate reader assays, we have already proven the functionality of our genetic constructs combined in one single strain. So, the next step was to demonstrate that the system also works if the constructs are separated in two different strains. Therefore, we continued with co-cultures of SeSt and ReSt using cell culture inserts (ThinCerts<sup>TM</sup>). Thereby, we could finally show that the SeSt stimulates the ReSt, containing a P<sub><i>srfA</i></sub>-<i>luxABCDE</i> fusion, by using luminescence as output (Figure 10). In the control, without SeSt, the <i>comX</i>-deficient ReSt exhibited only a weak luminescence signal because of the promoter’s basal activity. Hence, our system, consisting of SeSt and ReSt, is qualified as a proof of principle for communication between two distinct strains.</p> | ||
</figure> | </figure> | ||
| − | |||
<hr> | <hr> | ||
| − | <h3>Co-culture of SeSt and ReSt using | + | <h3 id="peptidosomes">Co-culture of SeSt and ReSt using Peptidosomes</h3> |
| + | |||
<p>The last step to an overall success of this subproject was the demonstration of microbial communication between bacteria encapsulated in Peptidosomes and bacteria in the surrounding medium. Therefore, we repeated the co-culture assay as described before but instead of separating the distinct strains using cell culture inserts we encapsulated the ReSt within a Peptidosome while we cultivated the SeSt in the surrounding medium. Luminescence was detected via well scan to precisely detect even small changes as well as the spatial extent. We could show that there is only a rise of luminescence if you cultivate the encapsulated ReSt together with the SeSt in the surrounding medium.</p> | <p>The last step to an overall success of this subproject was the demonstration of microbial communication between bacteria encapsulated in Peptidosomes and bacteria in the surrounding medium. Therefore, we repeated the co-culture assay as described before but instead of separating the distinct strains using cell culture inserts we encapsulated the ReSt within a Peptidosome while we cultivated the SeSt in the surrounding medium. Luminescence was detected via well scan to precisely detect even small changes as well as the spatial extent. We could show that there is only a rise of luminescence if you cultivate the encapsulated ReSt together with the SeSt in the surrounding medium.</p> | ||
| − | |||
| − | <figure class="makeresponsive" style=" | + | <figure> |
| + | <figure class="makeresponsive floatright" style="width:60%;" > | ||
<img src="https://static.igem.org/mediawiki/2017/0/04/T--TU_Dresden--P_Communication_Wellscan.png" | <img src="https://static.igem.org/mediawiki/2017/0/04/T--TU_Dresden--P_Communication_Wellscan.png" | ||
alt="Figure 11 Wellscan" class="zoom"> | alt="Figure 11 Wellscan" class="zoom"> | ||
| Line 261: | Line 276: | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | |||
| + | <p>In Figure 11 A, B and C illustrate the experimental set-up whereas D, E and F display the results of the well scan analysis. While areas with luminescence less or equal the maximal luminescence measured for SeSt (≤ 40 RLU) are blue-colored, areas with luminescence above SeSt level (≥ 40 RLU) are pink-colored. Therefore, we can show that the increase of luminescence is restricted to the Peptidosome (Figure 11, E). The mean luminesescence determined for the encapsulated ReSt is 90 RLU. There are few areas with values that are negligibly above the threshold (41 to 62 RLU) causing pink-colored squares located outside the expected position of the Peptidosome. This is a consequence of the chosen threshold and provides no evidence for cells leaving the Peptidosome. <b></p> | ||
| + | </figure> | ||
| + | |||
<hr> | <hr> | ||
<h2>Conclusion</h2> | <h2>Conclusion</h2> | ||
| − | <p class="survey-quote"=><b>Our team has made substantial progress in the evaluation of Peptidosomes as a tool for co-cultivation and studies of microbial interactions. We could prove communication between encapsulated bacteria and bacteria in the surroundings by exchanging | + | <p class="survey-quote"=><b>Our team has made substantial progress in the evaluation of Peptidosomes as a tool for co-cultivation and studies of microbial interactions. We could prove communication between encapsulated bacteria and bacteria in the surroundings by exchanging signaling molecules (=ComX). Further research about pore sizes of the Peptidosome membrane and diffusion rates needs to be done to tap the full potential of Peptidosomes as a powerful co-culture technique.</b></p> |
</div> | </div> | ||
Latest revision as of 14:48, 13 December 2017