Both of the below sections represent improved characterization of parts [http://parts.igem.org/Part:BBa_J04450 J04450], [http://parts.igem.org/Part:BBa_K1399010 K1399010], [http://parts.igem.org/Part:BBa_K1399011 K1399011], and [http://parts.igem.org/Part:BBa_K1399012 K1399012]
Expression of Red Fluorescence Protein in High Salt Media
As part of our experiments to determine if genetically modified E. coli can grow and thrive in salt water, we plated dilutions of E. coli exposed to different media on agar to obtain CFU/mL counts. We plated them not only on LB + chloramphenicol, but also LB with 3% NaCl (mimicking seawater conditions) to recover any other microorganisms that might be present in the local water mixtures we used. Surprisingly, the E. coli that grew on these plates seemed to not only grow slower, but express less RFP (a as evidenced by decreased fluorescence that was not recovered when given a longer incubation time).
We repeated this experiment by restreaking these bacteria, in addition to other variants of RFP (primarily to test another hypothesis, described below) that are degraded faster, on plates with 3% NaCl. We also tested these on plates that either did, or did not, contain IPTG - normally, no IPTG is necessary to observe fluorescence with construct BBa_J04450, since expression is apparently leaky enough.
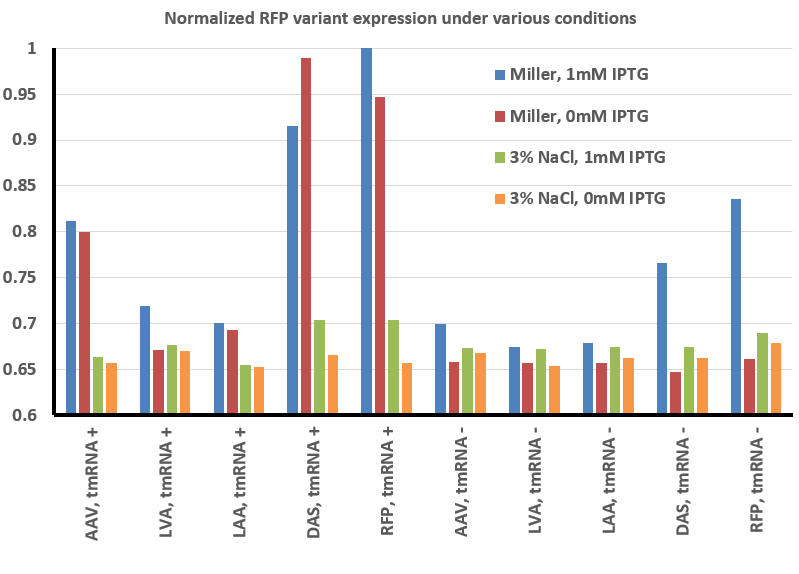
To summarize our results thus far, we have found that there is less fluorescence when grown in the presence of 3% NaCl with RFP and all the RFP variants we tested. In addition, expression appears to be controlled in a tighter fashion - whereas IPTG is not necessary to observe fluorescence under normal conditions, in the presence of high salt the expression is less 'leaky' and only observed when the gene is actually induced.
We argue that this is an important result for the iGEM community, as it brings with it a word of caution. Your gene circuit and design may work as intended under laboratory conditions, but how robust is it? Will something as simple as a shift in salt concentration change the parameters of your design? Will it be more robust? These are all important questions to answer.
Expression and Degradation of Red Fluorescence Protein variants in Δ ssrA cells
Our project was to create a kill-switch that will allow bacteria to survive under one set of conditions (darkness) but not another (light). In order to prevent premature cell death, we included an ssrA degradation tag to our kill-switch toxin protein, MazF. Interestingly, this may lead to a positive feedback effect: if enough MazF is produced, it will cleave mRNAs and create a lot of stalled ribosomes - this in turn results in a lot of other ssrA tagged proteins in the cell. Since the cell has a finite supply of proteases that recognize the MazF tag, the increase in ssrA tagged proteins should create a queuing effect at the protease, and indirectly stabilize MazF.
We wanted to test this idea, but to do so by testing the opposite scenario: in a cell that lacks tmRNA and any endogenous ssrA tagged proteins, would MazF-ssrA be degraded faster? As a stand in for MazF in this experiment, we used RFP - either untagged, or tagged with different variants of the ssrA tag. The normal ssrA tag, ANDENYALAA, is effectively recognized by proteases such as ClpXP and Lon. In constructs such as K1399012, it is represented by the last three amino acid letters (RFP-LAA); other constructs we tested, such as K1399010 (RFP-AAV), K1399011 (RFP-LVA), or K1399013 (RFP-DAS) have versions of the tag that are less efficiently recognized. We hoped that expression of these constructs in a cell that lacks tmRNA would lead them to be recognized better than they normally are. For example, bacteria with the RFP-DAS construct normally accumulate enough RFP to appear red on a LB plate - would growth in a ΔssrA cell diminish the amount of RFP to an extent that could be visualized on a plate?
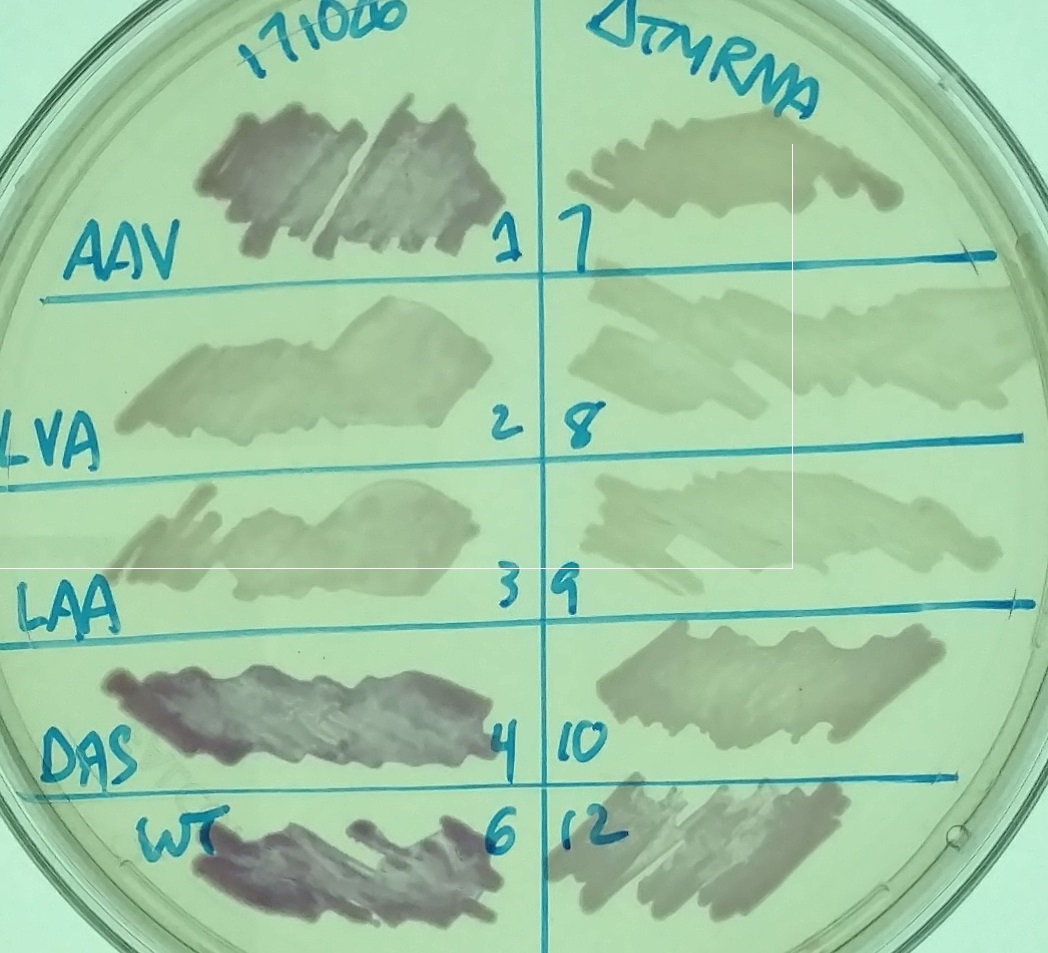
The image on the left shows a petri dish with LB Miller agar, chloramphenicol, and 1mM IPTG with various strains - different variants of RFP in strains that have (left) or lack (right) tmRNA. The labels refer to the different types of ssrA tags on the RFP.
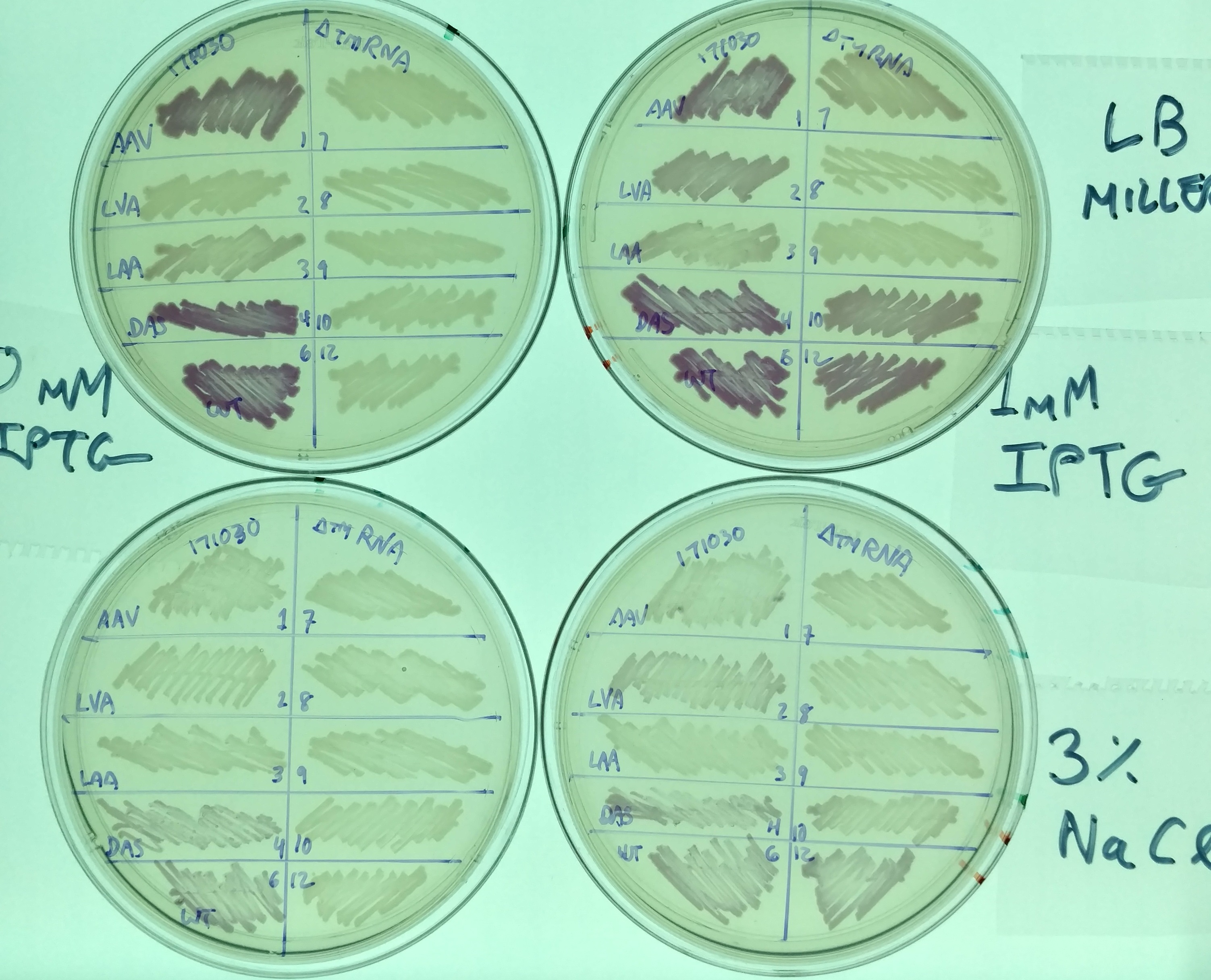
Another representative image illustrates the same effect, as well as the difference in expression in these variants between normal LB miller (with 1% NaCl - top two plates) and high salt media (LB supplemented with 3% NaCl - bottom two plates). It also illustrates the effect tmRNA has on how the lac promoter is regulated - without tmRNA, expression is less leaky. This is evident from comparing the plates on the left (No induction, 0mM IPTG) with those on the right (strong induction, 1mM IPTG). This result is consistent with previous observations (basically, that the lacI regulator is a normal target of tmRNA tagging and degradation), but illustrates the effect in a way not previously observed (i.e. was not thought to prevent leaky expression to this extent).
Our results suggest that the presence or absence of other ssrA tagged proteins does effect the degradation rate of a tagged reporter protein. Further experiments are necessary to confirm this result and rule out other explanations - for example, perhaps the decreased amount of RFP is simply reflective of a diminished capacity for translation in the cell. Complementing these cells with a tmRNA variant that adds a tag not recognized well by the proteases (such as tmRNA-DD or tmRNA-H6) would help test this idea.
The image above was a representative result, and was analyzed using ImageJ (see our Methods page for more details). The result of this analysis is shown below: