Survival of Genetically Modified E. coli in a Saline Environment
The 2016 Kingsborough iGEM team was focused on importing a new nitrogen metabolism pathway into E. coli; the anammox pathway. This pathway could potentially make E. coli more efficient at degrading the nitrogenous waste molecules found in sewage. The intended use of such a microbe would be in a wastewater treatment plant; however, this carries with it a risk that the bacteria could escape the confines of the facility and disrupt the local ecosystem.
Many iGEM teams address the risks of unintended spread of their modified organism by introducing (or at least proposing to introduce) a ‘kill-switch’. In fact, the focus of the Kingsborough team this year is to develop a new kill switch to be used in a wastewater treatment plant environment. However, we wanted to first assess the risk. Can a modified E. coli really survive outside the intended environment, and spread through the local waterways? Will it retain its plasmid and new genetic elements while doing so?
To answer these questions, we took a commonly used BioBrick, BBa_J04450, that expressed Red Fluorescent Protein as our stand-in for a genetic modification. This choice made it easy to track the presence of the plasmid, since the bacteria are colored red when they express these plasmid genes. We tested the ability of this bacteria to survive or thrive and maintain its plasmid multiple times.
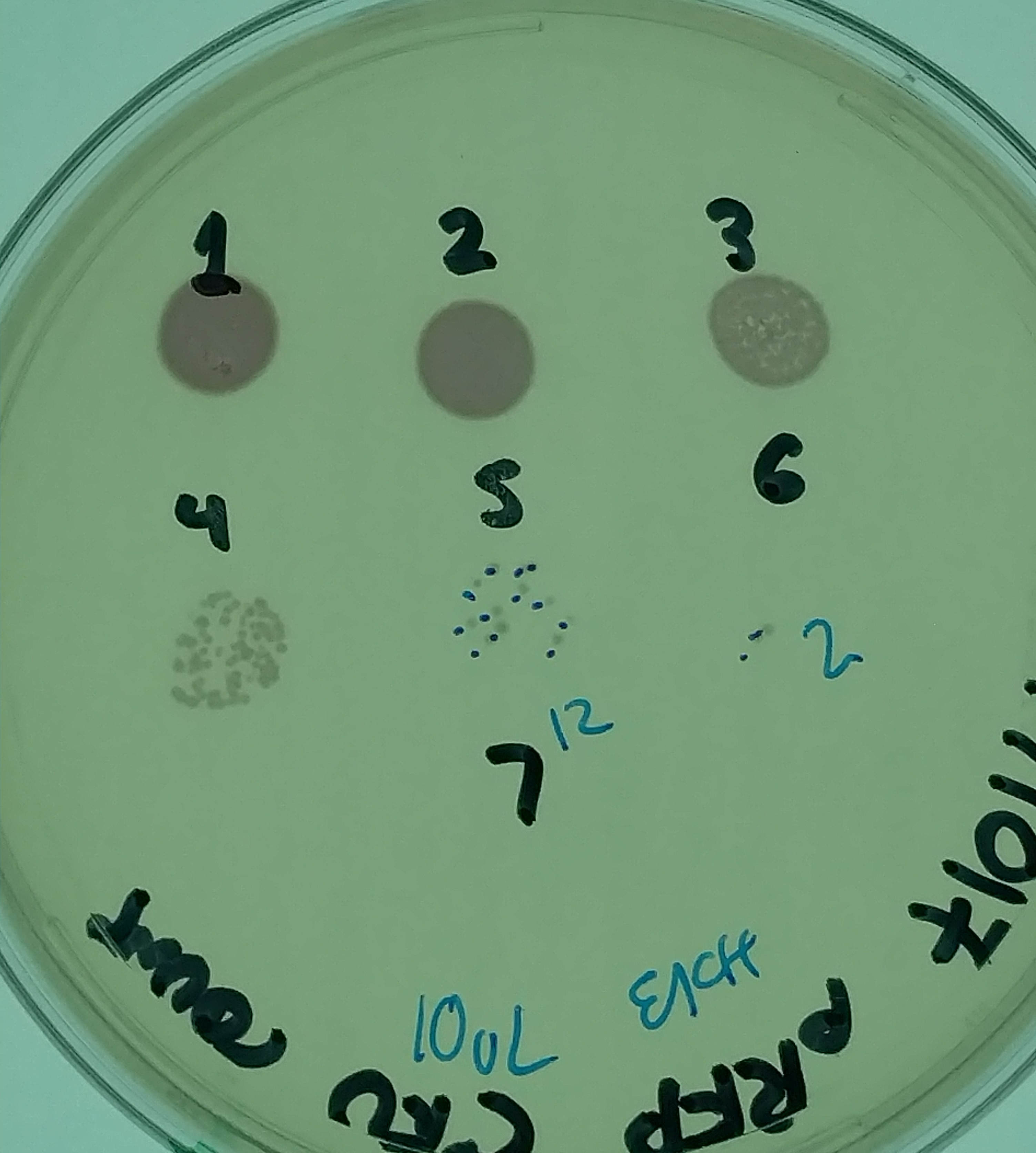
In one representative experiment, we started by growing a 5mL culture of E. coli with plasmid J04450 overnight in LB with chloramphenicol (Cm). 1mL of this culture was centrifuged, and the pellet was resuspended in sterile saline solution (0.9% NaCl). Dilutions of this mixture were then plated on LB + Cm by spotting 10uL of bacteria and allowing the drop to dry into the plate. This gave the following result (picture on the right):
This indicated that the starting suspension had about 1.6 x 10^8 CFU/mL of bacteria. 50uL of this mixture was then added to 5mL of LB, seawater (SW), or sterilized seawater (SSW), with or without an additional 500uL of LB being added (preliminary experiments added this volume of bacterial culture directly, without centrifugation). The amount of bacteria added, given their new volume, means that each culture should start out with 1.6 X 10^6 CFU/mL
The Results: E. coli can survive, but not thrive
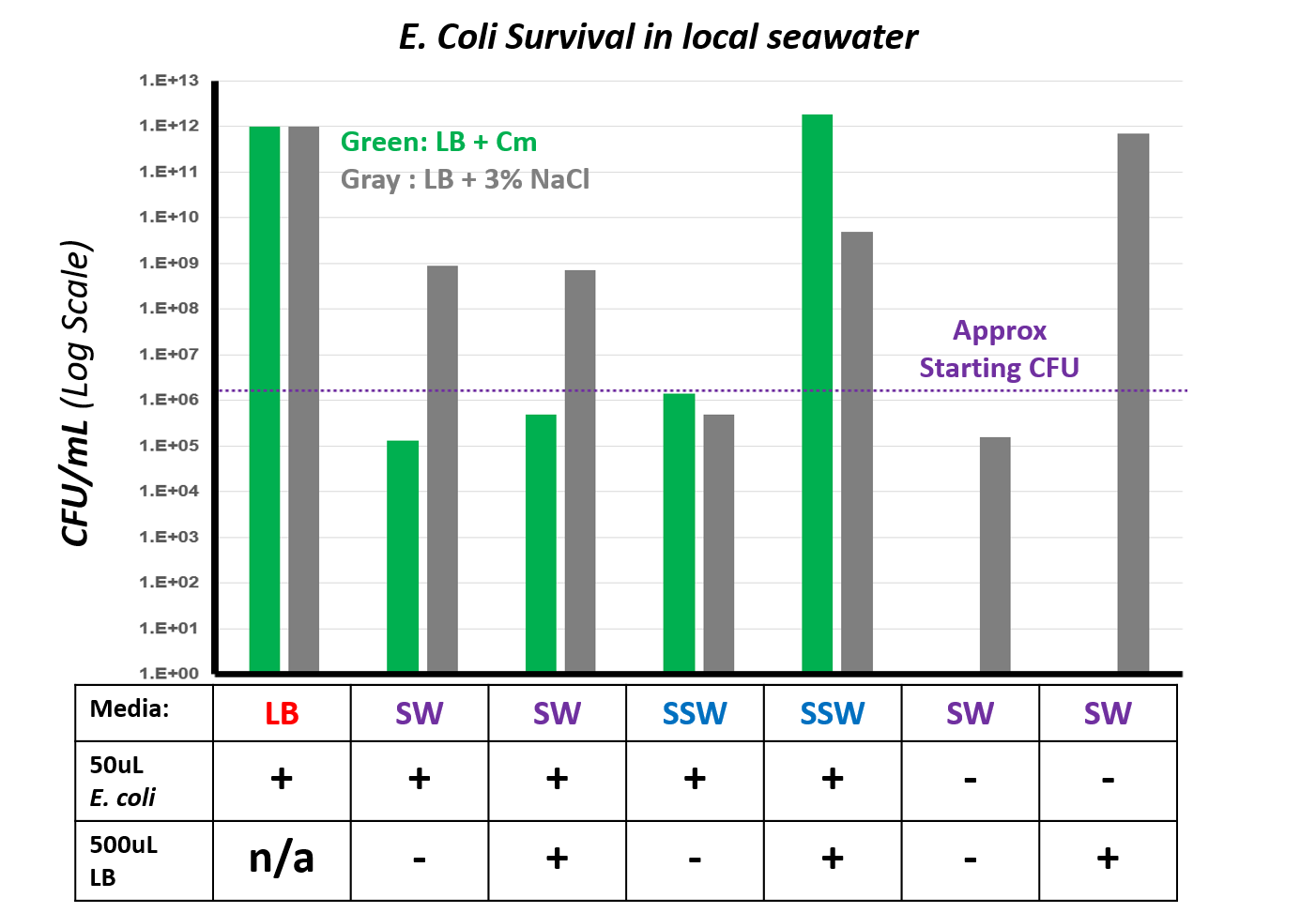
After allowing the bacteria to incubate overnight, we then spot-plated dilutions using the same protocol as before to determine bacterial density. Instead of just plating the cells on LB + Cm, we also plated them on LB without the selective antibiotic, but containing 3% NaCl to support the growth of organisms that might be found in the seawater. This way, we can determine if potential competitors are present and inhibiting E. coli growth. The resulting CFU/mL values are shown below – the dotted purple line indicates the starting amount of bacteria in each culture tube.
The results we obtained suggest a number of things:
- E. coli will maintain plasmid J04450 without the presence of the selective antibiotic. When grown in LB without Cm overnight, the number of Cm resistant bacteria grows from the initial 1.6x10^6 CFU/mL to over a million times that amount.
-
The abiotic stress of higher salt content in seawater does not phase E. coli. The bacteria grew almost as well in sterile seawater supplemented with additional nutrients as it did in LB.
- There are not that many nutrients usable by E. coli present in the seawater sample. Without the addition of 500uL of LB to the sterile seawater, E. coli was able to tolerate the conditions but not proliferate.
- Competition from other organisms or attack by bacteriophages limit E. coli numbers in seawater. Growth in unsterilized seawater was reduced compared to the number of bacteria initially seeded in the culture.
Another interesting result that came out of this experiment was that E. coli harboring part BBa_J04450, when grown on plates with 3% NaCl, did not produce as much Red Fluorescent Protein as bacteria grown on normal LB Miller (with 1% NaCl). These observations were tested more systematically in another set of experiments.