Djcamenares (Talk | contribs) |
Djcamenares (Talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Kingsborough_NY}} | {{Template:Kingsborough_NY}} | ||
| − | + | On this page you will find sections detailing the results we got on our journey for creating the BioBricks we submitted to the registry this year. | |
| + | |||
| + | Not shown are the many attempts necessary, and the failed creation of other BioBricks. For example, we were unsuccessful in creating BioBricks K2268005 and K2268006, but our failure (together with other data) suggests that the uninduced levels of MazF expression was simply too high and too great a threat to cell vitality. | ||
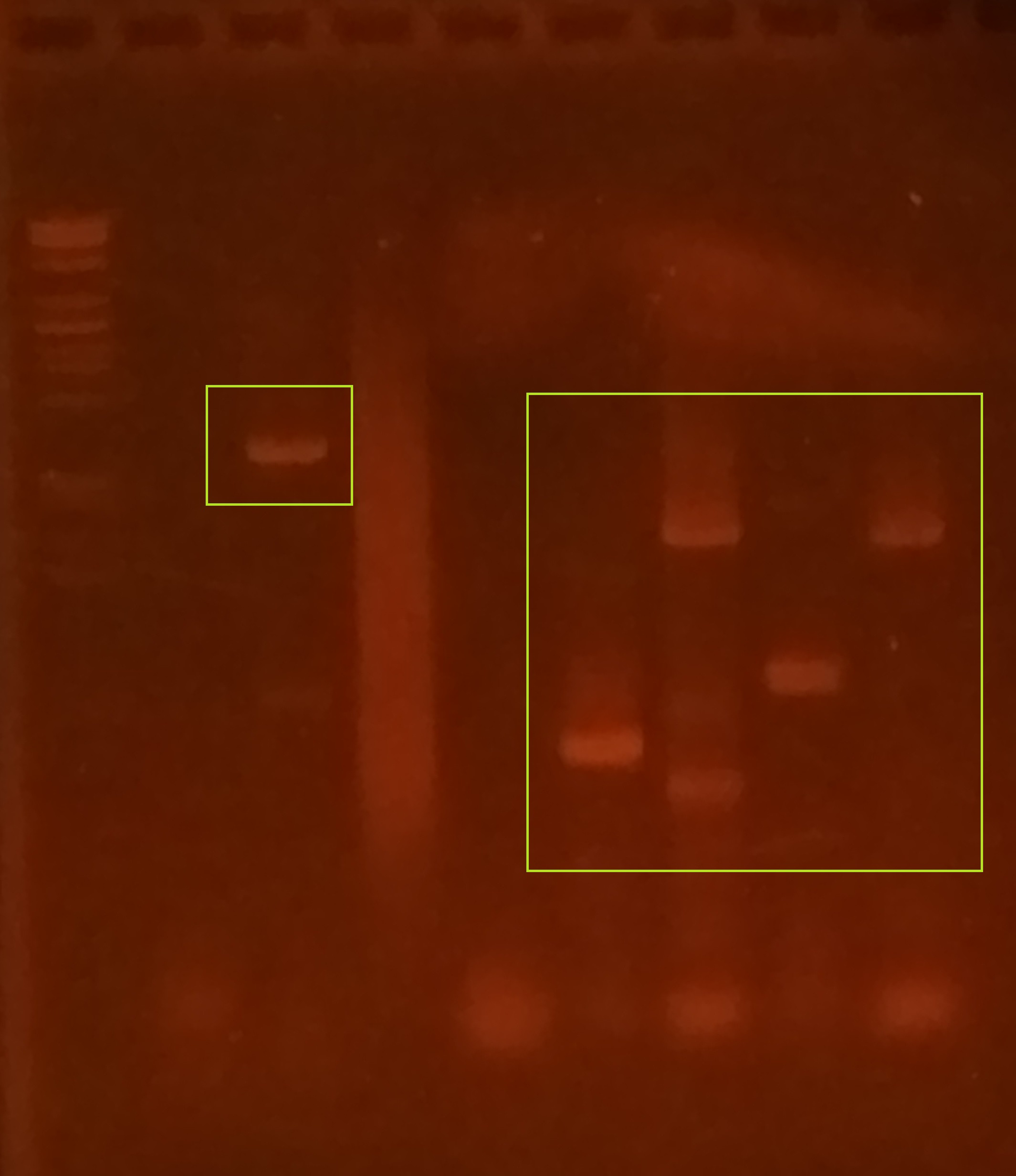
== PCR to Generate Fragments for Gibson Assembly == | == PCR to Generate Fragments for Gibson Assembly == | ||
| − | + | [[File:T--Kingsborough_NY--gibson_fragment_pcr1.jpg|600px|right]] | |
| + | |||
| + | Fragments obtained from IDT to construct our plasmid designs were not suitable for Gibson Assembly. Therefore, we ordered primers and performed PCR to generate fragments with homology to each other and/or the pSB1C3 plasmid backbone. These fragments were resolved on an agarose gel (shown on the right). | ||
| − | + | The first lane contains our molecular weight marker: Lambda DNA digested with EcoRI and PstI. the first small yellow box shows amplification of a single fragment containing all the genes for construct K2268006 (MazF and cI repressor). The second yellow box shows the various fragments (representing LacZ-alpha, cI repressor, or MazF-ssrA with appropriate homologies) that were subsequently used in Gibson Assembly (after purification, of course!). | |
<br clear=all> | <br clear=all> | ||
| Line 13: | Line 17: | ||
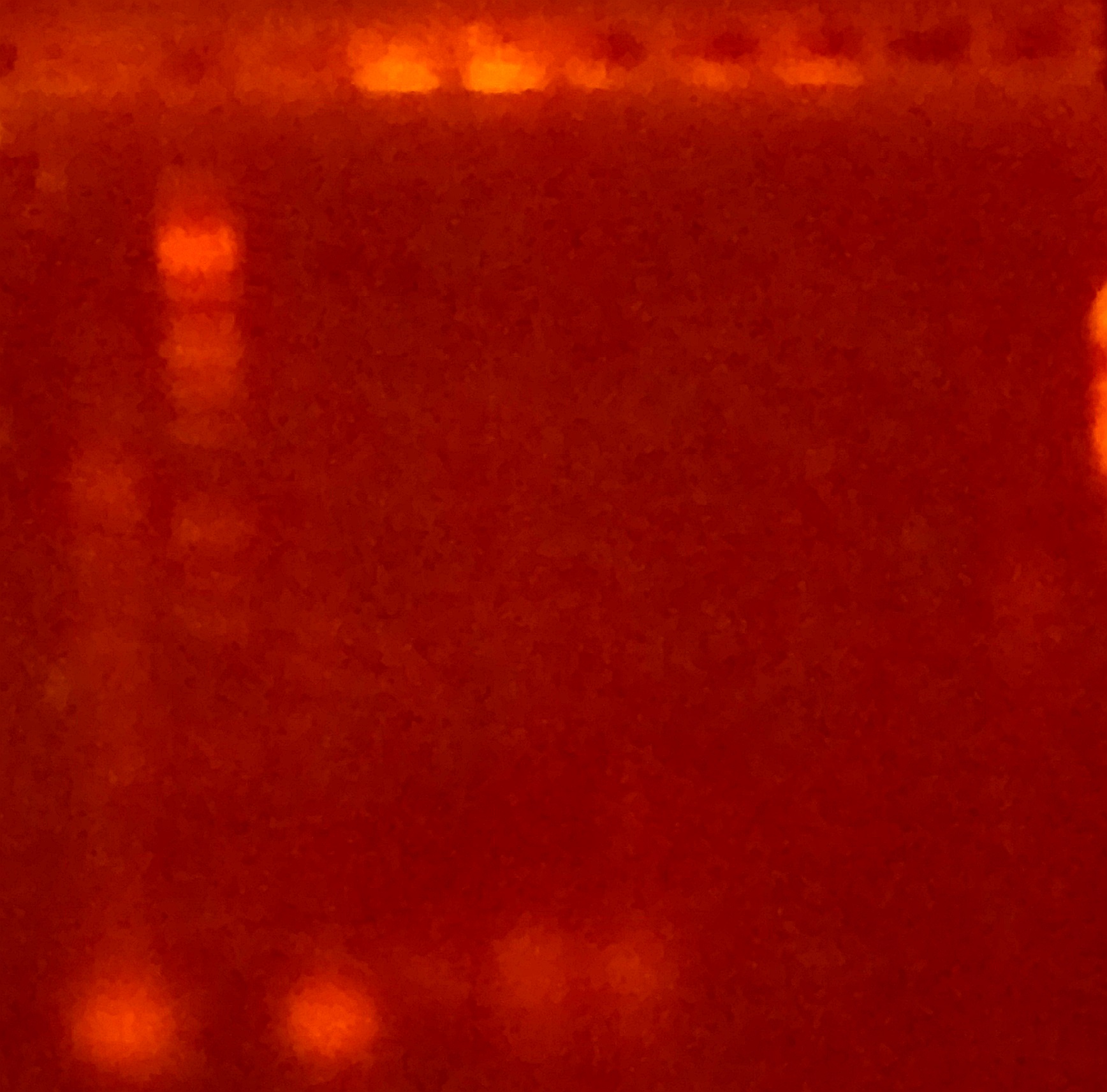
== Colony PCR to Verify Clone is Correct == | == Colony PCR to Verify Clone is Correct == | ||
| − | + | [[File:T--Kingsborough_NY--colony_pcr1.jpg|600px|right]] | |
| + | |||
| + | For construct K2268004, the Gibson Assembly was apparently successful, and multiple clones were obtained. The Gibson control reaction and transformation control also worked. These clones were amplified using the standard iGEM sequencing primers, VF2 and VR, which have homology to the pSB1C3 backbone. | ||
| + | |||
| + | The first lane contains a successful colony PCR, giving a band that corresponds in size to the genes of construct K2268004 (compare with similar band from previous section). The second band is our molecular weight marker, Lambda DNA digested with EcoRI and PstI (prepared in-house). | ||
| − | |||
<br clear=all> | <br clear=all> | ||
| Line 21: | Line 28: | ||
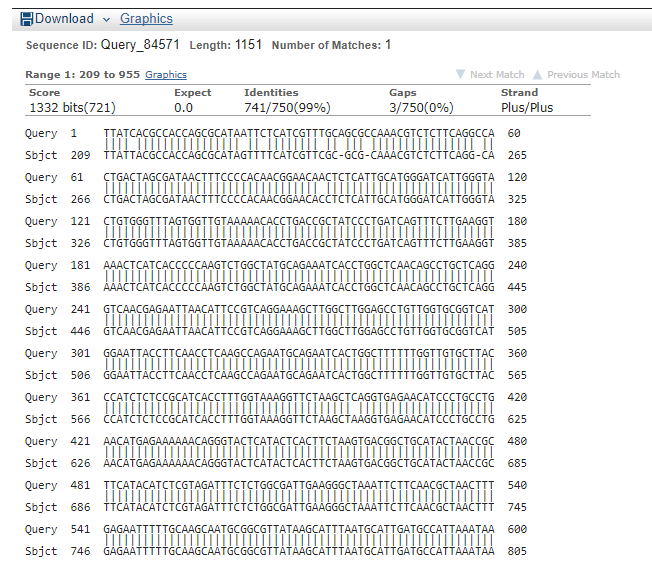
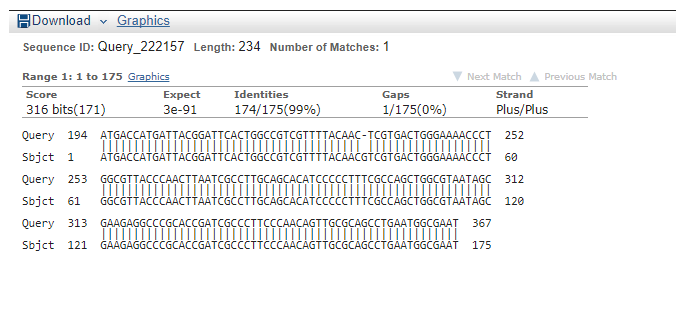
== Sequencing Verification == | == Sequencing Verification == | ||
| − | + | Multiple clones for different constructs were sent to be sequenced by [https://www.etonbio.com/ Eton Biosciences] using VF2, VR, other primers (see below). | |
| + | |||
| + | The images below show the result of a Blast-2-Seq analysis using the sequencing results for K2268004 (query) and either the cI repressor gene sequence (left) or the LacZ-alpha gene sequence (right). | ||
| + | <br clear=all> | ||
| + | |||
| + | [[File:T--Kingsborough_NY--sequencing_comparison_K2268004_cI.png|550px|left]] | ||
| + | |||
| + | [[File:T--Kingsborough_NY--sequencing_comparison_K2268004_lacZa.png|450px|right]] | ||
| + | |||
| + | <br clear=all> | ||
| + | |||
| + | Full sequencing results for construct K2268004: | ||
| − | + | <b> VF2 Primer result, reverse complement </b> | |
| − | > | + | <font face="Courier New"> |
GGGNACGTTACGTTGCGCATTTAACTATTAGATGCGTCGAATCACGAGGCAGGAATTTCAGATAAAAAAAATCCTTAGCT | GGGNACGTTACGTTGCGCATTTAACTATTAGATGCGTCGAATCACGAGGCAGGAATTTCAGATAAAAAAAATCCTTAGCT | ||
TTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTG | TTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTG | ||
| Line 31: | Line 49: | ||
TGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGC | TGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGC | ||
CCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAAT | CCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAAT | ||
| + | </font> | ||
| + | <b> 3.2_LinkR Primer result, reverse complement </b> | ||
| − | > | + | <font face="Courier New"> |
ATCACCAGCTCCCGTCTTTCATTGCATACGAAATTCCGGAGAGCATTCATCAGGCGGGCA | ATCACCAGCTCCCGTCTTTCATTGCATACGAAATTCCGGAGAGCATTCATCAGGCGGGCA | ||
AGAAATGTGAATAAAGGCCGAATAAAACTTGTGCTTATTTTTCTTTACGGTCTTTAAAAA | AGAAATGTGAATAAAGGCCGAATAAAACTTGTGCTTATTTTTCTTTACGGTCTTTAAAAA | ||
| Line 48: | Line 68: | ||
ACAGAGGGCAGCTTGAATGGCGAATGGGCTTTGCCTGGTTTTTCGAATTCTTCCGCAACT | ACAGAGGGCAGCTTGAATGGCGAATGGGCTTTGCCTGGTTTTTCGAATTCTTCCGCAACT | ||
TACTGCGTAGCGNA | TACTGCGTAGCGNA | ||
| + | </font> | ||
| − | > 3.2_LinkF Primer result | + | <b> 3.2_LinkF Primer result </b> |
| + | |||
| + | <font face="Courier New"> | ||
NTCCATGATAAGTACTCGCACTTTCTCAGCAGTCATCAGTTCGCCGCGCCAAACGTCTCTTCAGGCCACTGACTAGCGAT | NTCCATGATAAGTACTCGCACTTTCTCAGCAGTCATCAGTTCGCCGCGCCAAACGTCTCTTCAGGCCACTGACTAGCGAT | ||
AACTTTCCCCACAACGGAACAACTCTCATTGCATGGGATCATTGGGTACTGTGGGTTTAGTGGTTGTAAAAACACCTGAC | AACTTTCCCCACAACGGAACAACTCTCATTGCATGGGATCATTGGGTACTGTGGGTTTAGTGGTTGTAAAAACACCTGAC | ||
| Line 65: | Line 88: | ||
GCAGGACTCGTAAAAGCCGCGTGCTGGCGATTCACAAGGCTCGCCCCCTGACGGCACTACAAAATCGACGCTCTAGGCAG | GCAGGACTCGTAAAAGCCGCGTGCTGGCGATTCACAAGGCTCGCCCCCTGACGGCACTACAAAATCGACGCTCTAGGCAG | ||
AGGTGCGAACCGCACGGACATAAAGGATCCAGC | AGGTGCGAACCGCACGGACATAAAGGATCCAGC | ||
| + | </font> | ||
| − | > VR Primer result, reverse complement | + | <b>* VR Primer result, reverse complement</b> |
| + | |||
| + | <font face="Courier New"> | ||
ATGATTCGGATCCCTGGCCGTGTTTACACGTTCGGAACTGGGAAACCCTGCGTACCAACT | ATGATTCGGATCCCTGGCCGTGTTTACACGTTCGGAACTGGGAAACCCTGCGTACCAACT | ||
AATCCCTTGCAGCACATCCCTTTCGCAGCTGCGTAATAGCGAAGAGGCCGCCCGATCGCC | AATCCCTTGCAGCACATCCCTTTCGCAGCTGCGTAATAGCGAAGAGGCCGCCCGATCGCC | ||
| Line 88: | Line 114: | ||
GCGTATTGGGCGCTCTGCCCCTCACTCGCTCCCTCTGGAAACTGCGGCAGTACGTTGCGC | GCGTATTGGGCGCTCTGCCCCTCACTCGCTCCCTCTGGAAACTGCGGCAGTACGTTGCGC | ||
GGAATGTAAGG | GGAATGTAAGG | ||
| + | </font> | ||
| + | |||
| + | The sequencing primers that were used included the 3.2 Link F Primer (ggtgccggaaagctggctggagtaataaTtattacgccaccagcgcatag) and 3.2 Link R Primer (ctatgcgctggtggcgtaataAttattactccagccagctttccggcacc) | ||
<br clear=all> | <br clear=all> | ||
Latest revision as of 01:28, 2 November 2017
On this page you will find sections detailing the results we got on our journey for creating the BioBricks we submitted to the registry this year.
Not shown are the many attempts necessary, and the failed creation of other BioBricks. For example, we were unsuccessful in creating BioBricks K2268005 and K2268006, but our failure (together with other data) suggests that the uninduced levels of MazF expression was simply too high and too great a threat to cell vitality.
PCR to Generate Fragments for Gibson Assembly
Fragments obtained from IDT to construct our plasmid designs were not suitable for Gibson Assembly. Therefore, we ordered primers and performed PCR to generate fragments with homology to each other and/or the pSB1C3 plasmid backbone. These fragments were resolved on an agarose gel (shown on the right).
The first lane contains our molecular weight marker: Lambda DNA digested with EcoRI and PstI. the first small yellow box shows amplification of a single fragment containing all the genes for construct K2268006 (MazF and cI repressor). The second yellow box shows the various fragments (representing LacZ-alpha, cI repressor, or MazF-ssrA with appropriate homologies) that were subsequently used in Gibson Assembly (after purification, of course!).
Colony PCR to Verify Clone is Correct
For construct K2268004, the Gibson Assembly was apparently successful, and multiple clones were obtained. The Gibson control reaction and transformation control also worked. These clones were amplified using the standard iGEM sequencing primers, VF2 and VR, which have homology to the pSB1C3 backbone.
The first lane contains a successful colony PCR, giving a band that corresponds in size to the genes of construct K2268004 (compare with similar band from previous section). The second band is our molecular weight marker, Lambda DNA digested with EcoRI and PstI (prepared in-house).
Sequencing Verification
Multiple clones for different constructs were sent to be sequenced by Eton Biosciences using VF2, VR, other primers (see below).
The images below show the result of a Blast-2-Seq analysis using the sequencing results for K2268004 (query) and either the cI repressor gene sequence (left) or the LacZ-alpha gene sequence (right).
Full sequencing results for construct K2268004:
VF2 Primer result, reverse complement
GGGNACGTTACGTTGCGCATTTAACTATTAGATGCGTCGAATCACGAGGCAGGAATTTCAGATAAAAAAAATCCTTAGCT TTCGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTG ATAATGGTTGCGCTAGCAAAGAGGAGAAAGCTTATGACCATGATTACGGATTCACTGGCCGTCGTTTTACAACTCGTGAC TGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGC CCGCACCGATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAAT
3.2_LinkR Primer result, reverse complement
ATCACCAGCTCCCGTCTTTCATTGCATACGAAATTCCGGAGAGCATTCATCAGGCGGGCA AGAAATGTGAATAAAGGCCGAATAAAACTTGTGCTTATTTTTCTTTACGGTCTTTAAAAA GGCCGTAATATCCAACTGAACGGTCTGGTTATAGGTACATTGAGCACTGACTGAAATGCC CTCAAAATGTTCTTTACGATGCCATTGGGATATATCACCGGTGGTATATCCAGTGATTTT TTTCTCCATTTTAGCTTCCTTAGCTCCTGAAAATCTCGATAACTCAAAAAATACGCCCGG TAGTGATCTTATTTCATTATGGTGAAAGTTGGAACCTCTTACGTGCCCGATCAACTCGAG TGCCACCTGACGTCTAAGAACCCATTATTATCGTGACATTAACCTATAAAAATAGGCGTA TCACGAGGCAGAATTTCAGATAAAAAAAATCCTTAGCTTTCGCTAAGGATGATTTCTGGA ATTCGCGGCCGCTTCTAGAGTAACACCGTGCGTGTTGACTATTTTACCTCTGGCGGTGAT AATGGTTGCGCTAGCAAAGAGGAGAAAGCTTATGACCATGATTACGGATTCACTGGCCGT CGTTTTACAACGTCGTGACTGGGAAAACCCTGGCGTTACCCAACTTAATCGCCTTGCAGC ACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGGCCCGCACCGATCGCCCTTCCCA ACAGAGGGCAGCTTGAATGGCGAATGGGCTTTGCCTGGTTTTTCGAATTCTTCCGCAACT TACTGCGTAGCGNA
3.2_LinkF Primer result
NTCCATGATAAGTACTCGCACTTTCTCAGCAGTCATCAGTTCGCCGCGCCAAACGTCTCTTCAGGCCACTGACTAGCGAT AACTTTCCCCACAACGGAACAACTCTCATTGCATGGGATCATTGGGTACTGTGGGTTTAGTGGTTGTAAAAACACCTGAC CGCTATCCCTGATCAGTTTCTTGAAGGTAAACTCATCACCCCCAAGTCTGGCTATGCAGAAATCACCTGGCTCAACAGCC TGCTCAGGGTCAACGAGAATTAACATTCCGTCAGGAAAGCTTGGCTTGGAGCCTGTTGGTGCGGTCATGGAATTACCTTC AACCTCAAGCCAGAATGCAGAATCACTGGCTTTTTTGGTTGTGCTTACCCATCTCTCCGCATCACCTTTGGTAAAGGTTC TAAGCTAAGGTGAGAACATCCCTGCCTGAACATGAGAAAAAACAGGGTACTCATACTCACTTCTAAGTGACGGCTGCATA CTAACCGCTTCATACATCTCGTAGATTTCTCTGGCGATTGAAGGGCTAAATTCTTCAACGCTAACTTTGAGAATTTTTGC AAGCAATGCGGCGTTATAAGCATTTAATGCATTGATGCCATTAAATAAAGCACCAACGCCTGACTGCCCCATCCCCATCT TGTCTGCGACAGATTCCTGGGATAAGCCAAGTTCATTTTTCTTTTTTTCATAAATTGCTTTAAGGCGACGTGCGTCCTCA AGCTGCTCTTGTGTTAATGGTTTCTTTTTTGTGCTCATAAGCTTTCTTCCTCTTTGCTAGCTGTGCTCAGTATCTTGTTA TCCGCTCACAATTTACTAGTAGCGGCCGCTGCAGGCCCGGCAAAAAAGGCAAGGGGTCACCACCCTGCCCTTTTTCTTTA AAACCGAAAAGATTACTTCGCGTTATGCAGGGTTCCTCGCTCACTGACTCGCTGCGCTCGGGCGTCGGCTGCGGCGGAGC GGATCAGCTCACTCAAGCGGTAATACGGTATCACAGAATCAGGGATACCGCAGAAGAACATGTGAGCAAAGGCAGCAAAG GCAGGACTCGTAAAAGCCGCGTGCTGGCGATTCACAAGGCTCGCCCCCTGACGGCACTACAAAATCGACGCTCTAGGCAG AGGTGCGAACCGCACGGACATAAAGGATCCAGC
* VR Primer result, reverse complement
ATGATTCGGATCCCTGGCCGTGTTTACACGTTCGGAACTGGGAAACCCTGCGTACCAACT AATCCCTTGCAGCACATCCCTTTCGCAGCTGCGTAATAGCGAAGAGGCCGCCCGATCGCC CTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGCGCTTGCCTGGTTTCCGGCACCAGAA GCGGTGCCGAAAGCTGGCTGGAGTAATATTATTACGCCACCAGCGCATAGTTTTCATCGT TCGCGCGCAAACGTCTCTTCAGGCACTGACTAGCGATAACTTTCCCCACAACGGAACACC TCTCATTGCATGGGATCATTGGGTACTGTGGGTTTAGTGGTTGTAAAAACACCTGACCGC TATCCCTGATCAGTTTCTTGAAGGTAAACTCATCACCCCCAAGTCTGGCTATGCAGAAAT CACCTGGCTCAACAGCCTGCTCAGGGTCAACGAGAATTAACATTCCGTCAGGAAAGCTTG GCTTGGAGCCTGTTGGTGCGGTCATGGAATTACCTTCAACCTCAAGCCAGAATGCAGAAT CACTGGCTTTTTTGGTTGTGCTTACCCATCTCTCCGCATCACCTTTGGTAAAGGTTCTAA GCTAAGGTGAGAACATCCCTGCCTGAACATGAGAAAAAACAGGGTACTCATACTCACTTC TAAGTGACGGCTGCATACTAACCGCTTCATACATCTCGTAGATTTCTCTGGCGATTGAAG GGCTAAATTCTTCAACGCTAACTTTGAGAATTTTTGCAAGCAATGCGGCGTTATAAGCAT TTAATGCATTGATGCCATTAAATAAAGCACCAACGCCTGACTGCCCCATCCCCATCTTGT CTGCGACAGATTCCTGGGATAAGCCAAGTTCATTTTTCTTTTTTTCATAAATTGCTTTAA GGCGACGTGCGTCCTCAAGCTGCTCTTGTGTTAATGGTTTCTTTTTTGTGCTCATAAGCG TTAGTATAACATTGAAGAGCTATTCGCATCACACGGATCTCCGAGCGCATTGAGCCATTG AACGAGGAATCGGAAGAGCGCCCATTAATCAATCGCCCCTCGCGCGGGGAGAGGCGGTTT GCGTATTGGGCGCTCTGCCCCTCACTCGCTCCCTCTGGAAACTGCGGCAGTACGTTGCGC GGAATGTAAGG
The sequencing primers that were used included the 3.2 Link F Primer (ggtgccggaaagctggctggagtaataaTtattacgccaccagcgcatag) and 3.2 Link R Primer (ctatgcgctggtggcgtaataAttattactccagccagctttccggcacc)