Below you can find a summary of the protocols we most often used in our experiments. They will allow you to better analyze our results, and hopefully will inspire some creative experimental methods in your own lab!
Contents
Bacterial Transformation
We utilized the common "TSS" transformation protocol:
Briefly, 1X TSS solution was prepared by mixing 0.5g of Polyethylene glycol (PEG) 3350 with 250uL of Dimethyl Sulfoxide (DMSO), 150uL of 1M MgCl2, and 4.5mL of sterile LB. This mixture was dissolved, and then filter sterilized (stored at 4C for future use).
Approximately mid-log bacteria were pelleted by centrifuging 15 mins at 3500 rpm in the cold (4C). The supernatant was removed, and the cells were resuspended with TSS at 1/10 their original volume.
Then 2-4ul of DNA was added to 100uL of cells, the cells were incubated on ice for 30 minutes, heat shocked for 30 seconds at 42C, and placed back on ice for 2 minutes. 600uL of sterile LB was added and the cells recovered for 60 mins at 37C. 100ul of the mixture was spread onto LB plates with the appropriate selective antibiotics.
For some of our constructs, we expected the cells to be light sensitive - if exposed to too much light, they may express a toxin which would prevent growth of the transformed cells. Thus, when performing these transformations, we carried out the experiments in the dark!
Molecular Biology
For Gibson assembly, we used the NEB HiFi DNA Assembly Kit. The fragments used were obtained from IDT and/or prepared via PCR reactions using the Q5 Master Mix from New England Biolabs (NEB). Restriction Enzymes were also obtained from NEB, and we used the Fast-Ligation kit from Bio-Basic. For routine PCR reactions (such as diagnostic colony PCR), we used a PCR kit from Bio-Basic.
DNA Quantification
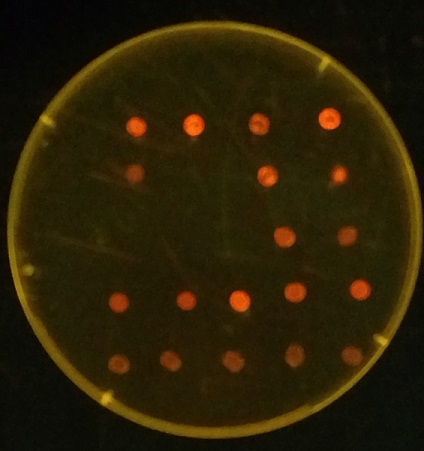
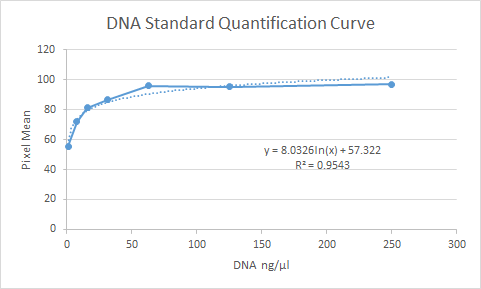
To quantify DNA and determine the concentration, we used an Ethidium Bromide (EtBr) Spot test. A spot of 10uL of dilute EtBr (1ng/uL) was mixed with 1uL of a DNA sample, placed over an UV lamp, and photographed. By using standards such as diluted lambda DNA at known concentrations, we generated a standard curve that allowed us to estimate the DNA in a sample. It might not have the precision of a nanodrop, but it uses the same amount of sample and is a lot less expensive! See a representative image to the right:
This protocol was adapted from the following paper: Kasap, et al (2006) Ethidium Bromide Spot Test Is a Simple Yet Highly Accurate Method in Determining DNA Concentration. Turk. J. Med. Sci 36 (6): 383-386
The formula on the standard curve can be interpreted as such:
Y values: DNA concentration
X Value: Pixel Intensity
b (Constant): Pixel Intensity with no DNA
Preparation of Media
LB Miller broth and plates were prepared by dissolving 12.5g of [http://www.labdepotinc.com/p-58630-lb-broth-miller-s.php LB power] alone (broth) or with 7.5g of [http://www.labdepotinc.com/p-58610-gc-agar.php Agar] base (plates) into 500mL of distilled H20. The mixture was then autoclave sterilized.
Concentrated 1000X stocks of antibiotics, such as Ampicillin (Am), Kanamycin (Kn), or Chloramphenicol (Cm) were used to supplement media appropriately, and were prepared as follows:
| Antibiotic | 1X Conc. | Grams | Solvent (10mL) |
|---|---|---|---|
| Am | 100ug/mL | 1.0g | dH20 |
| Kn | 50ug/mL | 0.5g | dH20 |
| Cm | 30ug/mL | 0.3g | 70% EtOH |
X-gal was prepared as a 1000X stock solution of 20mg/mL; 0.2g was added to 10mL of DMF, and stored protected from light at -20C.
IPTG was prepared as a 1000X stock solution of 1M; 2.38g was added to 10mL of distilled H20 (dH20), filter sterilized, and stored at 4C in the short-term or divided into aliquots and stored at -20C.
Since LB Miller normally contains 1% NaCl, higher salt plates were generated by adding either 10g of NaCl (for a total of 3% NaCl) or 20g of NaCl (for a total of 5% NaCl) to the 500mL LB-Agar mixture listed above.
Agarose Gel Electrophoresis
To separate out different sized DNA fragments, we prepared 1% agarose gels using the 0.5g EZ Agarose tablets dissolved in 50mL of 10mM NaBO4 buffer. The same buffer was used to run the gel once loaded with samples, typically at a range of 120 - 150 volts. After sufficient run time, the gel was removed, mixed with approximately 50mL of distilled water and 5uL of Ethidium Bromide (at a 10mg/mL stock concentration) for 10 minutes, and then destained with fresh distilled water for about 5 minutes before visualizing on a UV lamp.
RFP and LacZ-Alpha quantification
How the color of bacterial growth on a petri dish is quantified:
1. Open a file in ImageJ (example file to try is given below)
2. Using the circle/oval or rectangle selection tool, select a small area of bacterial growth that is more or less uniform in color. (i.e. don't select a region that contains both bacteria and empty / blank plate.)
3. Under the Analyze menu, select Histogram (or hit CTRL+H). You should see a graph with one peak - if there are multiple large peaks, go back to step 2 and try to get a more uniform selection.
4. The default measurement is grayscale. Record the average grayscale (the mean) value for that selection, then click on RGB to cycle to a value that represents the amount of red in the selection. Record this mean as well, and cycle through green and blue, recording each value for that selection.
5. Repeat the process above for all other bacteria growths on the plate, recording their values separately, and 1 extra set of measurements for a blank spot on the plate (a negative control). It may be easier to move to the next bacterial growth by selecting the "Live" option in the histogram window and drawing your selection shape to the part of the picture with the next bacterial growth.
6. Calculate the ratio between the red and green values (for RFP), or blue and red values (for LacZ-alpha) for any given bacterial growth, and subtract from it any background signal (i.e. the plate itself). This ratio approximates the amount of pigment produced in the bacteria.
Seawater Experiments
For experiments testing E. coli survival in seawater, local water samples were obtained from a dock on the north end of campus, which is part of the opening to Sheepshead Bay (which filters into Rockaway Inlet and then into the Atlantic Ocean.) The date and time of each sample was noted to compare it with tide levels, temperature, and other environmental variables.
500uL of a culture of E. coli (containing BioBrick BBa_J04450), or sterile LB, or an E. coli pellet re-suspended in sterile saline solution, was added to tubes containing 5mL of the seawater sample (SW) or the same sample after autoclave sterilization (sterilized seawater, SSW). The cultures were allowed to incubate overnight in a tube rotor.
Bacterial density in cultures were determined by creating 1:10 dilutions in sterile saline solution and plating these dilutions (for example, spotting 10uL of each dilution) onto appropriate LB plates. Colonies counted the next day were multiples by the dilution factor (# of 1:10 dilutions x 100) to determine CFU/mL count.