| Line 300: | Line 300: | ||
<h3 class="team_name">Filippo Abbondanza</h3> | <h3 class="team_name">Filippo Abbondanza</h3> | ||
<p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | <p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Developing the lysoenic lambda phage carrying the FnCpf1 CRISPR system.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | <a href=" | + | <a href="https://www.facebook.com/filippo.abbondanza" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 316: | Line 313: | ||
<h3 class="team_name">Erin Corbett</h3> | <h3 class="team_name">Erin Corbett</h3> | ||
<p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | <p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Engineering E. coli to create our mock pathogen testing platform, and engineering the lytic T7 phage.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
</p> | </p> | ||
</div> | </div> | ||
| Line 332: | Line 325: | ||
<h3 class="team_name">Rikki He</h3> | <h3 class="team_name">Rikki He</h3> | ||
<p class="team_designation">MSc Biochemistry</p> | <p class="team_designation">MSc Biochemistry</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Developing the lysogenic P1 phage carrying spCas9 CRISPR system.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | <a href=" | + | <a href="https://www.facebook.com/y.q.heybd" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 347: | Line 338: | ||
<h3 class="team_name">Ti He</h3> | <h3 class="team_name">Ti He</h3> | ||
<p class="team_designation">MSc Biotechnology</p> | <p class="team_designation">MSc Biotechnology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Developing the lysogenic lambda phage carring saCas9 CRISPR system.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | <a href=" | + | <a href="https://www.facebook.com/profile.php?id=100012772305809" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 365: | Line 353: | ||
<h3 class="team_name">Lydia Mapstone</h3> | <h3 class="team_name">Lydia Mapstone</h3> | ||
<p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | <p class="team_designation">MSc Synthetic Biology & Biotechnology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Engineering E. coli to create our mock pathogen testing platform and engineering the lytic T4 phage using BRED.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | |||
| − | |||
| − | |||
| − | |||
</p> | </p> | ||
</div> | </div> | ||
| Line 381: | Line 365: | ||
<h3 class="team_name">Yuri Matsueda</h3> | <h3 class="team_name">Yuri Matsueda</h3> | ||
<p class="team_designation">MSc Biotechnology</p> | <p class="team_designation">MSc Biotechnology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Engineering P1 lysogenic phage with CRISPR-SaCas9 system to target resistance gene fragments.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | <a href=" | + | <a href="https://www.facebook.com/profile.php?id=100004212933551" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</div> | </div> | ||
| Line 397: | Line 378: | ||
<h3 class="team_name">Anton Puzorjov</h3> | <h3 class="team_name">Anton Puzorjov</h3> | ||
<p class="team_designation">MSc Bioinformatics</p> | <p class="team_designation">MSc Bioinformatics</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Building a model of bacteria-phage interactions in two-step re-sensitisation combining both lysogenic and lytic phages.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
<a href="https://www.facebook.com/puzorjov" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> | <a href="https://www.facebook.com/puzorjov" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> | ||
| Line 412: | Line 393: | ||
<h3 class="team_name">Yating Wang</h3> | <h3 class="team_name">Yating Wang</h3> | ||
<p class="team_designation">MSc Drug Discovery & Translational Biology</p> | <p class="team_designation">MSc Drug Discovery & Translational Biology</p> | ||
| − | <p class="team_text"> | + | <p class="team_text">Engineering P1 lysogenic phage with CRISPR-Cpf1 system to target resistance gene fragments.</p> |
<p class="social-icons"> | <p class="social-icons"> | ||
| − | <a href=" | + | <a href="https://www.facebook.com/profile.php?id=100010921308790" class="facebook" target="_blank"><i class="ion-social-facebook-outline"></i></a> |
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
</div> | </div> | ||
Revision as of 13:41, 12 June 2017
Antibiotic revolution: a molecular toolkit to re-sensitise
Methycillin resistant S.aureus (MRSA)
Vancomycin resistant S.aureus (VRSA)
Klebsiella pneumoniae
Pseudomonas aeruginosa
Enterococcus faecalis/faecium
Acinetobacter baumanii
Modular molecular toolkit for re-sensitisation of antibiotic-resistant pathogens using CRISPR delivered by a two-phage system.
Read moreABOUT PhagED
The world is currently facing a future where antibiotics no longer work and basic infections cannot be treated. Many of the most dangerous diseases which are not treatable arise from the hospital environment. This is because the bacteria have had lots of exposure to antibiotics, allowing them time to evolve genes which can help them survive in the presence of these drugs. We therefore want to tackle this problem of the 'super bugs' found in our hospitals, otherwise known as the ESKAPE pathogens.
We will do this by developing a spray which will contain modified phages. When that phage enters the cell, CRISPR-Cas system will cut up the genes which are giving the bacteria resistance to antibiotics. We then add another phage to ensure this system is maintainable.
By using this spray, we hope to re-sensitise these dangerous antibiotic resistant bacteria to the drugs again, allowing conventional antiobiotics to work. This would mean we would no longer need to develop costly new antibiotics, and avoid the coming doom of a world where a UTI is a death sentence. Neat eh?!
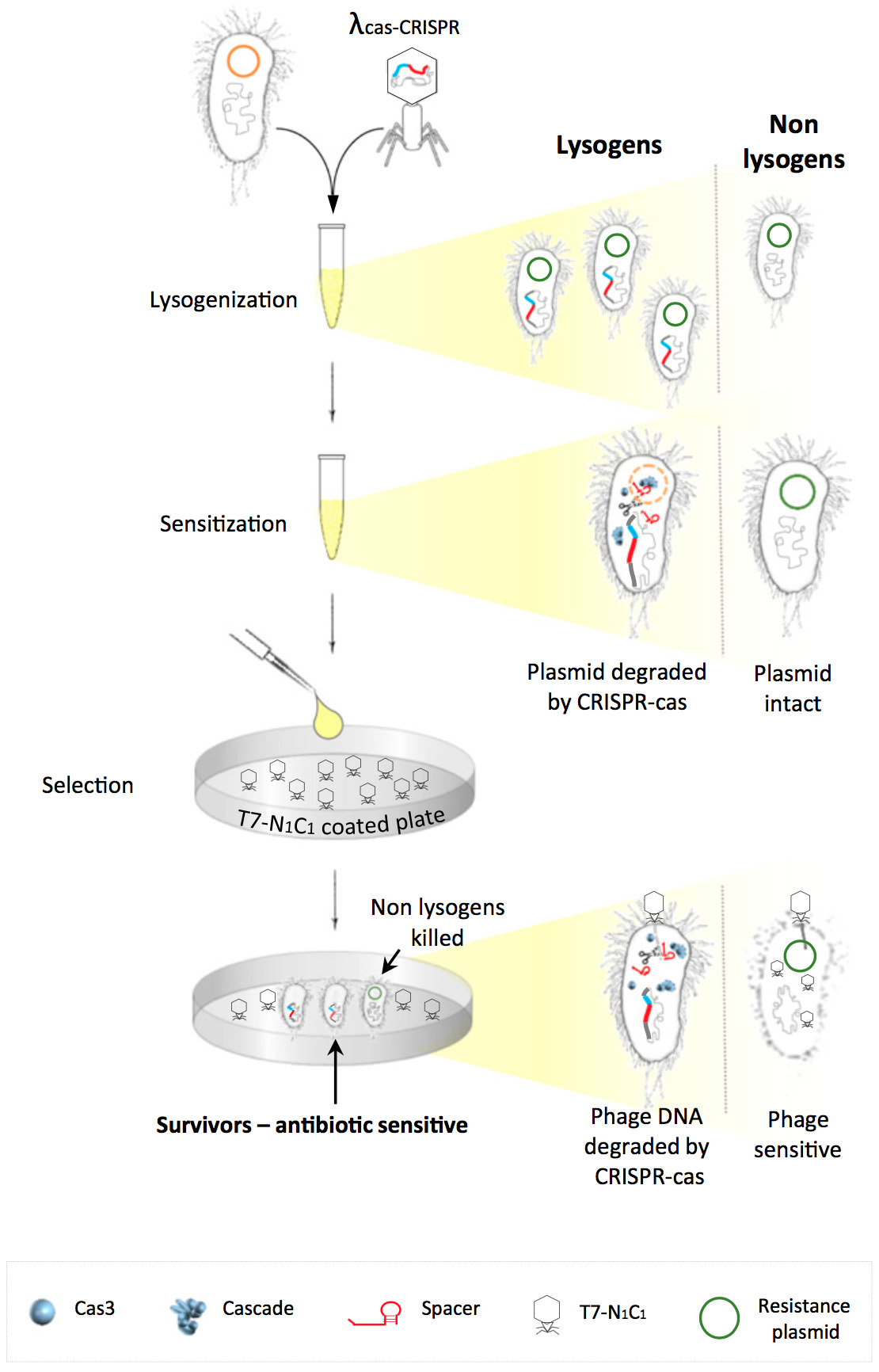
TAKE A CLOSER LOOK
Aliquam lobortis. Maecenas vestibulum mollis diam. Pellentesque auctor neque nec urna. Nulla sit amet est. Aenean posuere
tortor sed cursus feugiat, nunc augue blandit nunc, eu sollicitudin urna dolor sagittis lacus.

Project
Get to know PhagED better.
Parts
Blocks of DNA that we've used.

Safety
Safety always comes first.

Human Practices
How PhaED fits into the context.
Meet The Team

Filippo Abbondanza
MSc Synthetic Biology & Biotechnology
Developing the lysoenic lambda phage carrying the FnCpf1 CRISPR system.

Erin Corbett
MSc Synthetic Biology & Biotechnology
Engineering E. coli to create our mock pathogen testing platform, and engineering the lytic T7 phage.

Rikki He
MSc Biochemistry
Developing the lysogenic P1 phage carrying spCas9 CRISPR system.

Ti He
MSc Biotechnology
Developing the lysogenic lambda phage carring saCas9 CRISPR system.

Lydia Mapstone
MSc Synthetic Biology & Biotechnology
Engineering E. coli to create our mock pathogen testing platform and engineering the lytic T4 phage using BRED.

Yuri Matsueda
MSc Biotechnology
Engineering P1 lysogenic phage with CRISPR-SaCas9 system to target resistance gene fragments.

Anton Puzorjov
MSc Bioinformatics
Building a model of bacteria-phage interactions in two-step re-sensitisation combining both lysogenic and lytic phages.

Yating Wang
MSc Drug Discovery & Translational Biology
Engineering P1 lysogenic phage with CRISPR-Cpf1 system to target resistance gene fragments.

Owen Yeung
MSc Synthetic Biology & Biotechnology
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore
DO YOU WANT TO HELP US TURN IT INTO REALITY?
Feel free to drop us a line or two and we'll be in touch soon!
Contact

