Contents
Overview
The aim of our work is to develop a low-cost device to introduce the sample, extract the biomarker, xylulose, and house the biosensing bacteria, all in a self-contained device.
Xylulose extraction has been carried out previously in “Structural Analysis of the Capsular Polysaccharide from Campylobacter jejuni” (Gilbert et al., 2007) [1] using the fact that it is very acid labile. The process involved heating the sample with acetic acid to a temperature of 100°C. To enable the device to be portable and used in the field, we targeted the use of microfluidics techniques to extract and detect xylulose.
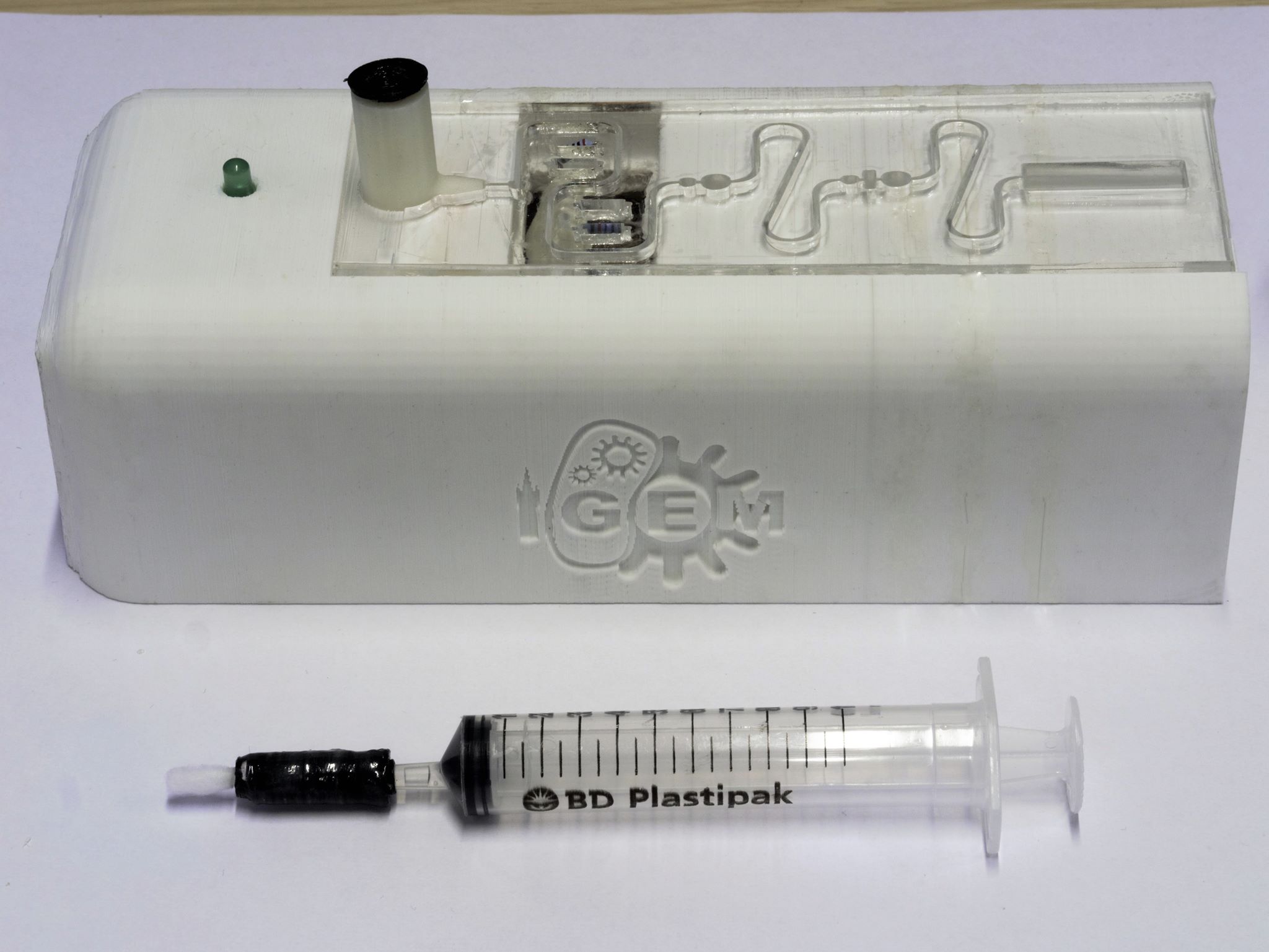
The device consists of 3 elements (as shown in figures X, Y and Z):
- A swab, used to gather the sample from the environment, attached to a syringe containing 1% acetic acid in tris acetate (TAE) buffer.
- A self-contained (to avoid contamination) combined processing plate and heating circuit for preparing the sample for detection.
- A Device casing to house and contain the processing plate and electronics
The processing plate contained a section to house the genetically modified bacteria that has been developed for our project, and allow the user to observe if there was any Campylobacter present in the original sample.
One part that is not available in this design is the detection system. In this version, we rely on eye inspection, but this could be implemented in a quantifiable system using for example a smartphone.
Challenges
- Neutralisation: Using acid for the extraction of xylulose would be detrimental to our biosensor down the line, killing our biosensing E.Coli. We thus have to neutralise the acid using an equivalent alkali. We also integrate a measurement of the pH to validate that the process is not harmful to our biosensor.
- Hierarchical Metabolism: Bacteria are able to metabolise xylulose [ref], which means that our biosensor carries the risk of not producing the reporter molecule upon addition of xylulose. We thus chose to add glucose to the sample before the contact with the biosensor, as E.Coli will metabolise glucose first, while xylulose is used by the biosensing pathway.
- Gel Encapsulation: Biosensor storage in the device. Our device is targeted at Point-of-need application and thus requires to be portable and to have all reagents already stored within the device.
- Heater Circuit: In order to carry out the heating step of the procedure, it was necessary to raise the temperature to 100°C. In light of this, a heater circuit was designed.
Final Design
A brief overview of how the device, shown in figure 1, is operated and the steps involved in processing the sample is given below:
A swab of the area for testing is taken, holding the sample. It is then inserted into the input section of the processing plate. The syringe, holding the solution of acetic acid, is pushed slowly to detach the sample from the swab matrix and pass it through the device. The sample is then taken through a series of stages which will release the sugar xylulose from the capsule of any campylobacter bacteria present. These being:
- Heating the acid and sample to 100 °C (which detaches xylulose)
- Neutralising the acid using a Tris-Acetate buffer (making the solution harmless to E. coli)
- Validating the pH using litmus paper
- Mixing with glucose (ensuring the E. coli does not metabolise the xylulose)
The sample is then delivered to the detection chamber, which holds a strip of filter paper with a hydrogel coating that contains our genetically modified E.coli cells. The detection chamber, along with the rest of the device, is transparent, allowing the user to observe any colour changes should xylulose be present in the sample.
A demonstrational video showing the finished device and outlining how it should be operated is given below in video 1.
<centre><video width="320" height="240" controls><source src="https://static.igem.org/mediawiki/2017/0/0a/T-Glasgow-Demonstrational_Video.mp4" type="video/mp4"></video></centre>
Condition set up
Sample preparation
- 1
- 2
- 3
Results and Discussion
Outlook
References
- ↑ Gilbert, M., Mandrell, R., Parker, C., Li, J. and Vinogradov, E. (2007). Structural Analysis of the Capsular Polysaccharide fromCampylobacter jejuni RM1221. ChemBioChem, 8(6), pp.625-631.