Contents
Overview
In the design of our Campylobacter biosensor we decided to employ a genetic logic circuit. This would require the presence of two transcriptional input signals from Campylobacter-associated molecules before an output signal in the form of GFP fluorescence could be produced. We aimed to build and characterise two genetic AND-gates; one based on a split-GFP fluorescence system; and the other based on a splitting the Enterobacteria phage T7 RNA polymerase (T7 RNAP). We wished to quantify the responsiveness of both AND-gates with well-characterised small-molecule regulated promoters from the iGEM registry, and then proceed to utilising our own Campylobacter sensing transcriptional systems to drive the AND-gate biosensor. Both split-GFP and split-T7 RNAP gates were constructed from BioBrick parts and tested, however DNA sequence errors appear to have prevented the proper function of either system.
Introduction
After brainstorming our Campylobacter biosensor project idea, we began investigating any molecules that are present within - or secreted by – Campylobacter. In an ideal scenario, we would detect a small molecule that is associated with gastroenteritis-causing Campylobacter only - not in any other non-pathogenic subspecies of the Campylobacter genus – nor present in any other bacteria that might be inhabit a poultry-carcass environment. Additionally, in this ideal scenario, a well-characterised E. coli gene regulatory system with a strong output expression level would already exist.
Sensation
We identified two molecules of interest from Campylobacter for detection. One was xylulose, a 5-carbon ketopentose sugar. The second was the quorum sensing molecule autoinducer-2. Xylulose is a rare sugar most commonly associated with the pentosephosphate pathway, where xylulose-5-phosphate is an intermediate step. Interestingly, xylulose was found in the polysaccharide capsule of Campylobacter jejuni strain RM1221 [1]. The presence of xylulose is not common in bacterial polysaccharide capsules, and the glyosidic bonds which incorporate xylulose were found to be extremely acid-labile. One sub-project aimed to exploit a xylulose-associated transcriptional activator system (mtlR-mtlE) from Pseudomonas fluorescens. Another subproject aimed to mutagenize the well-characterised arabinose transcriptional regulatory system (araC-pBAD) from E. coli and modify its ligand specificity to the closely related structure of xylulose. Our engineering subproject designed a functional prototype biosensor device that simplified a process of dissolving a swabbed input sample, processing the solution through acid and high temperature to release xylulose, and then delivering the sample to a waiting strain of genetically modified bacterial carrying the biosensor circuit for detection. Finally, purified xylulose is extremely expensive to purchase commercially, so for the testing of our biosensor we attempted to biosynthesise and purify the sugar ourselves. The other molecule we identified as a marker for Campylobacter was autoinducer-2 (AI-2), a secreted quorum sensing molecule. AI-2 is a significantly less specific biomarker, as many varied gram-positive and gram-negative bacterial species sense their population density and surrounding bacterial environment using this molecule [2]. On the other hand, this ubiquity meant that AI-2 gene regulation was well characterised, with prior iGEM teams having worked on the natural E. coli AI-2 quorum sensing regulatory system.
The AND-gate
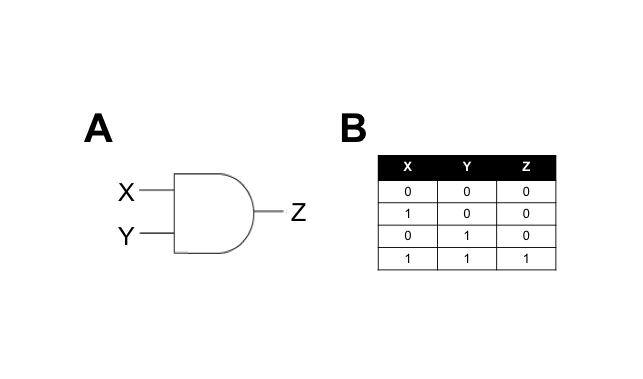
We hypothesised that building a combination biosensor requiring the input of two Campylobacter-associated molecules would reduce the risk of false-positives. To do this we envisaged a biosensor genetic circuit in the form of an AND-gate. In electronic circuitry, an AND-gate is a form of switch that only activates an output in the presence of two positive input signals (Figure 1).
A genetic AND-gate uses transcriptional signalling in place of electricity. In the simplest form, we can design a genetic circuit where two gene products must be actively transcribed to produce an output signal. The interaction between the input and output signals can be direct or indirect, and we aim to test both types in this subproject.
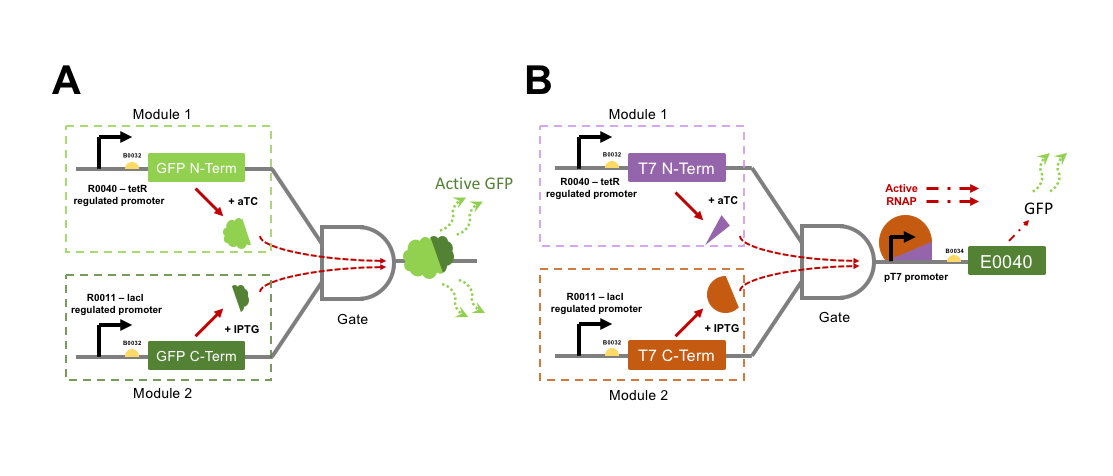
The directly interacting genetic AND-gate we aim to test is gained by GFP into two individual protein subunits, either of which expressed alone would not be fluorescent. If both GFP subunits were to be transcriptionally controlled from separate promoters within the same cell, then an AND-gate would be formed. Only expression of both subunits at the same time would make the cells fluorescent. To split GFP we attempted to improve parts designed by a previous iGEM team, Davidson Missouri 2007, who split the GFPmut3b part E0040 into a 151 aa N-terminal fragment (named GFP1: [http://parts.igem.org/Part:BBa_I715019 I715019]) and an 83 aa C-terminal fragment (named GFP2: [http://parts.igem.org/Part:BBa_I715020 I715020]). These parts were designed for fusion to other proteins for Bimolecular fluorescence complementation (BiFC) assays for validation of protein-protein interactions. As such, the N-terminal part lacked a stop codon, and the C-terminal part lacked a start codon. The 2015 team NUDT_CHINA altered the GFP1 part to include a stop codon, now labelled [http://parts.igem.org/Part:BBa_K1789003 K1789003], but we would have to design a PCR amplification to add a start codon to the GFP2 part for our uses.
For an indirectly interacting genetic AND-gate the inputs do not themselves act together as the output. The indirect gate we planned can be generated by splitting the RNA polymerase (RNAP) protein from Enterobacteria phage T7. T7 RNAP is a single subunit RNA polymerase that is most commonly used for strictly-controlled recombinant protein expression in E. coli. T7 RNAP begins transcription only from its cognate pT7 promoter sequence, and has complete orthogonality (no cross-reactivity) with E. coli promoter sequences. In addition, the T7 RNAP produces extremely reactive gene expression responses. Three hours following induction of T7 RNAP expression, a gene product expressed from the T7 promoter (pT7) may represent 50% of the cell’s mass [3]. Interestingly, a proportion of the purified T7 RNAP protein is often found cleaved into two subunits, one small (179 aa), and one large subunit (701 aa). If these two cleavage products are expressed as separate polypeptides within a cell then they are capable of driving gene expression from a pT7 promoter, although at a moderately lower efficiency than the wild-type single polypeptide protein [4]. A Split T7 RNAP genetic circuit represents an indirect AND-gate when each subunit is controlled by a separate transcriptional activator and the pT7 promoter drives expression of an output such a GFP. No previous iGEM team has attempted to split and test a T7 RNAP AND-gate, although many have utilised pT7 ([http://parts.igem.org/Part:BBa_K1321338 K1321338]) and T7 RNAP ([http://parts.igem.org/Part:BBa_I716103 I716103]) for protein over-expression.
We wished to build constructs of both split-GFP and split-T7 RNAP AND-gates by controlling the expression of split subunits from well-characterised orthogonal small-molecule regulated promoters within E. coli. Figure 2 shows a genetic diagram of our plan for testing the two different AND-gate circuits. It was an aim to quantify the difference in output GFP fluorescence level between the directly interactive split-GFP AND-gate, and the indirect T7-gate. If time was to permit, then testing would proceed to swapping input control of the gates over to the xylulose and autoinducer-2 regulated systems. Herein we will refer to each promoter + split protein construct as a “module”, i.e. a full AND-gate will require the presence and activation of both relevant modules.
Methods
Standard molecular biology techniques.
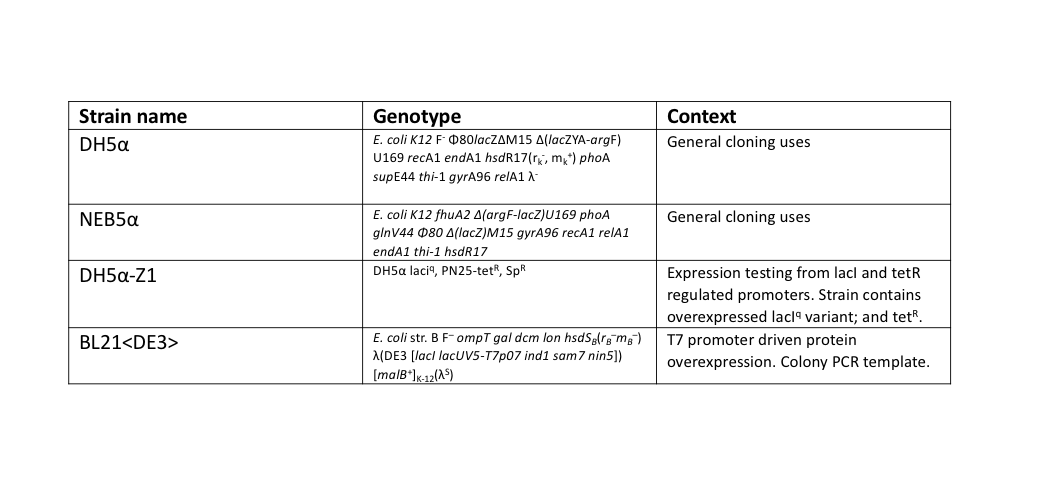
During this work, a standardised set for routine laboratory techniques were used such as restriction digest, gel electrophoresis, preparation of chemically competent E. coli, transformation of chemically competent E. coli, ligation reactions, oligonucleotide annealing, and PCR. These can be found on our protocols page. All ligation reactions described herein were transformed into NEB5α commercial chemically competent E. coli cells, plated on L-agar containing the required antibiotics, and grown at 37°C overnight.
PCR design
Split T7 RNA polymerase: PCR primers were designed to split T7 RNAP into two separate BioBrick compatible parts designed according to Shis and Bennett (2013)[4]. In their work, the 880 amino acid protein was split into 179 aa N-terminal, and a 701 aa C-terminal fragments. Forward and reverse primers to generate these two fragments were designed using Primer3 [5], aiming for primer annealing temperatures of roughly 60°C, and are shown in Table 2.
Primer design included the insertion of the weak strength ribosome binding site (RBS) B0032 upstream of the start codon of the two T7 RNAP fragments (green) highlighted sequence above. The addition of these RBS emulated the effect of BioBrick assembly by inclusion of the TACTAG assembly scar between the B0032 and RNAP fragment sequences (orange). Both fragment reverse primers were designed to result in the parts ending with double TAA stop codons, in the case of the N-term part this meant the insertion of the two stops, while the C-terminal part primer added one in addition to the natural TAA (red). The C-terminal F-primer also included a new start codon (purple). Forward primers included BioBrick prefixes and reverse primers included the BioBrick suffixes respectively, highlighted in yellow. All primers included a 6-bp sequence of DNA (blue) to allow efficient restriction digest of the outermost restriction enzymes, according to NEB instructions.
The T7 RNAP fragments were amplified by a colony PCR of E. coli BL21<DE3>, a strain of E. coli that has the T7 RNAP integrated into its chromosome. A single colony of BL21<DE3> was picked from a streaked agar plate, then resuspended into 200 μl of TE buffer. 1 μl of this colony template was used in 50 μl NEB Phusion polymerase reactions, following their protocol, except with an extended (10 min) initial 95°C denaturation step to release genomic DNA from the colony template.
Split GFPmut3b (E0040)
We designed a PCR reaction to generate a C-terminal GFP2 part which would work alongside the existing N-terminal GFP1 part K1789003 by NUDT_CHINA 2015. Primer3 was used to design roughly 60°C Tm primers to amplify the coding sequence of the 83 aa C-terminal fragment, as shown in Table 3.
The GFP2 part was designed without the addition of a RBS. Only a start codon (purple) was inserted into the part sequence. BioBrick prefix/suffix (yellow), and flanking DNA (blue) are included in the fragment amplification. A 1 in 1000 dilution of miniprep DNA of the full GFP part E0040 was used as template for an NEB Phusion polymerase reaction, following their PCR condition protocol.
SDS-PAGE
SDS-PAGE was performed using Bio-Rad 4–15% Mini-PROTEAN® TGX™ 10-well Precast Protein Gels, run in 1x Tris-Glycine buffer. E. coli GFP expression samples were prepared by recording the optical density of 1 ml samples of cells during culture, at 600 nm using a spectrophotometer. The 1 ml cell samples were transferred to separate microcentrifuge tubes and pelleted at 16,000 g for 5 minutes. Supernatant was discarded, and the cell pellets were resuspended in 2x final concentration Laemmli sample buffer (65.8 mM Tris-HCl, pH 6.8, 26.3% (w/v) glycerol, 2.1% SDS, 0.01% bromophenol blue, 20 mM DTT). Volume of sample-buffer to be added was normalised to OD values, where the highest OD sample received 100 μl of buffer, and lower ODs received less proportionally. Samples were boiled at 95°C for 5 minutes, then microcentrifuged at 16,000 g for 5 minutes. 20 μl of prepared samples were loaded into the pre-cast gel wells. Empty wells were filled with sample buffer. Gel-electrophoresis was performed at 150 V for roughly 45 minutes, or until the dye front reached the base of the gel. The gel was stained in Coomassie Blue (0.1% Coomassie Brilliant Blue G250, 50% methanol, 10% acetic acid) for 1 hour, then de-stained overnight in 10% methanol 10% acetic acid solution.
Results
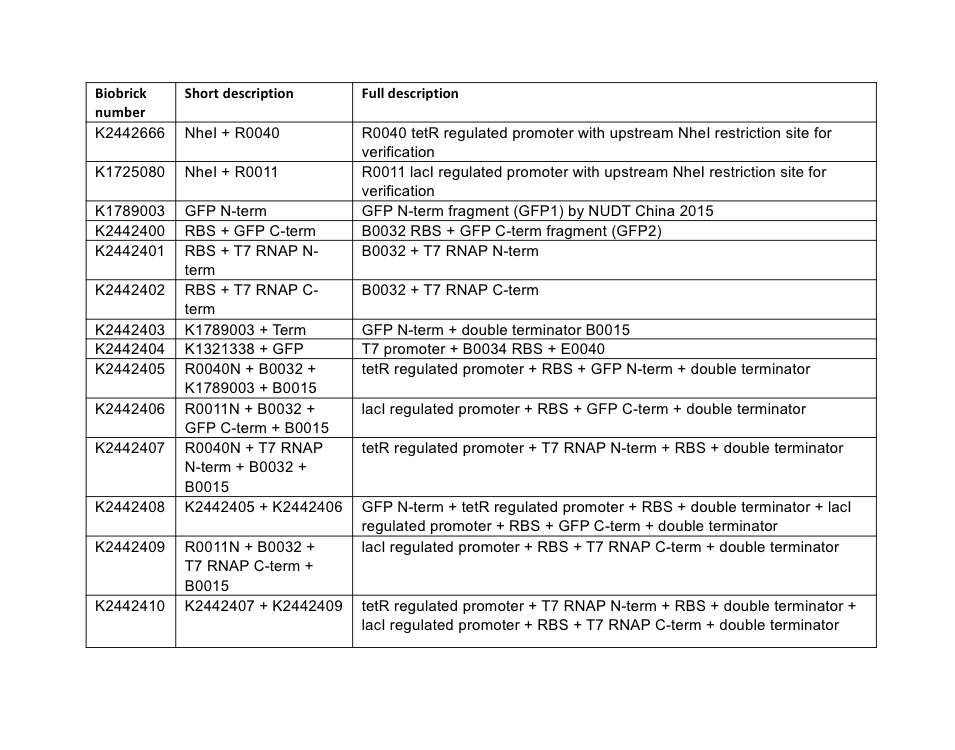
Plasmid assembly. All plasmids were assembled using BioBrick assembly techniques, and a full list of parts used in this subproject is described below in Table 4. All parts are assembled the standard BioBrick vector pSB1C3 unless otherwise stated.
Basic parts
PCR reactions were designed to amplify the C-terminal fragment of GFP, and both the N and C-terminal fragments of T7 RNAP. These three amplifications additionally inserted the weak ribosome binding site B0032 upstream of the coding sequence of these fragments. Following the reactions, gel electrophoresis confirmed successful amplification of the fragment parts. The fragments were purified following the QIAGEN PCR Cleanup kit protocol, then each was restriction digested for ligation into pSB1C3 vectors. Following transformation of the ligation reactions, three transformant colonies were picked, cultured overnight, and miniprepped. Minipreps were then checked by restriction digest for the presence of correct insert and vector sizes, which was confirmed correct for all three split-fragment parts. As a final confirmatory step, the GFP C-terminal, T7 N-term, and T7 C-term fragment BioBrick parts, now in the pSB1C3 vector, were sequenced using the VF2 BioBrick sequencing primer. Sequencing confirmed the correct assembly of the two T7 RNAP parts, but revealed an issue with the GFP C-terminal part – the PCR design had incorporated an incorrect version of the BioBrick prefix, shown in Figure 3.
As explained on the http://parts.igem.org/Help:BioBrick_Prefix_and_Suffix page, BioBrick parts beginning with ATG must use a shortened version of the prefix ending TAG, instead of the longer version for non-ATG parts which ends TAGAG. The short version ensures ideal spacing between upstream ribosome binding site parts and the start codon. Unfortunately, this oversight of the primer design was not initially noticed our plasmid map sequence for the C-terminal GFP part also contained the error. A part such as this with the extra two nucleotides before the start codon has the potential to be translated at decreased level from an upstream ribosome binding site.
AND-gate modules
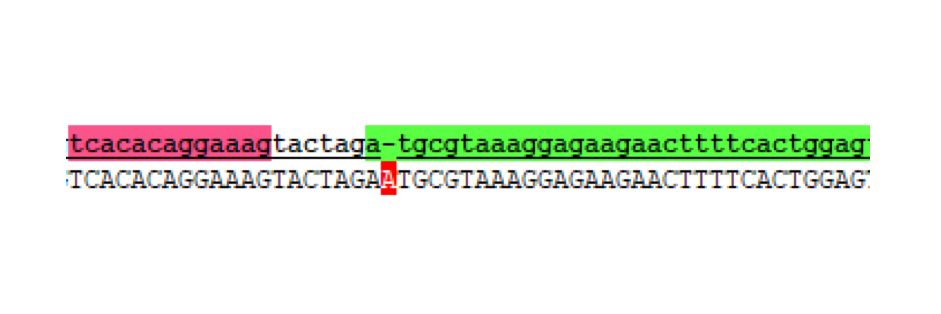
Without realising the sequence issue of the GFP C-terminal part, assembly proceeded for making the respective split fragments into individual AND-gate modules. The first design choice implemented for all four AND-gate modules was the inclusion of the double terminator part [http://parts.igem.org/Part:BBa_B0015 B0015]. This terminator part would be present at the end of each discrete module, and prevent read-through transcription from the preceding promoter through to the 2nd AND-gate module. The three split fragments created by PCR along with the pre-existing GFP N-terminal part K1789003 were restriction digested for insertion upstream of the B0015 terminator in pSB1C3. Following ligation, transformation, and restriction digest verification, these parts were sequenced. Both T7 RNAP modules were shown to be correct; however, at this stage the problem with the C-terminal GFP part was noticed, and a similar problem was discovered in the pre-existing N-terminal GFP part. The N-term GFP part contains an extra A nucleotide immediately preceding the start-codon ATG (Figure 4).
This erroneous nucleotide is present in the parts-registry sequencing for K1789003, but is not reported as an error. Unfortunately, now both split-GFP modules might be affected by improperly spaced ribosome binding sites. Despite this we proceeded to the next assembly stage.
A general design was chosen for both types of AND-gate, where one module of each gate would be placed under transcriptional control of a lacI-regulated promoter, and other module would be under control of a tetR-regulated promoter. The lacI ([http://parts.igem.org/Part:BBa_R0011 R0011]) and tetR ([http://parts.igem.org/Part:BBa_R0040 R0040]) regulated promoters were chosen to control the AND-gate modules as they are heavily characterised and well understood parts. We utilised a variant of both promoters (lacI: [http://parts.igem.org/Part:BBa_K1725080 K1725080], tetR: [http://parts.igem.org/Part:BBa_K2442666 K2442666]) where a NheI restriction site is inserted prior to the promoter sequence; this doesn’t alter the behaviour of the promoter but allows for an easy analytical restriction digest to verify the ligation of annealed promoter oligonucleotides. These promoters are simply known as R0011N and R0040N respectively.
At this stage, the three AND-gate modules that originated from PCR possessed a B0032 RBS, split-fragment coding sequence, and B0015 terminator. These modules required addition of promoters. The GFP N-terminal was only flanked by B0015, and required the addition of a promoter and ribosome binding site. Promoter insertion was done by using annealed oligonucleotides; this is a technique where the promoter sequence is ordered in the form of Forward and Reverse strand single-stranded oligonucleotides, where the BioBrick prefix and suffix have been truncated so that upon annealing a double-stranded promoter fragment is formed with BioBrick-compatible sticky-ends for ligation upstream of existing parts. The C-terminal module of both AND-gates received R0011N tetR-regulated annealed promoter oligos, and the C-terminal module of T7 RNAP gate received the R0040N lacI-regulated annealed promoter. The C-term module of split-GFP gate received a R0011+B0032 annealed oligo sequence. These annealed oligos were ligated upstream of the existing module parts by digests of the BioBrick prefix, then transformed into E. coli, minipreps verified by restriction digest, then sequenced using VF2 primer. Sequencing revealed that both split-GFP modules had assembled correctly, although maintained the pre-existing prefix errors; the C-terminal split T7 RNAP was also confirmed. However, the N-terminal T7 RNAP module was shown to possess a four-nucleotide deletion in its R0040N promoter (Figure 5).
Unfortunately, this sequencing alignment was overlooked at the time, and the deletion within the functional elements of the tetR-regulated promoter was not noticed until after the final assembly stage.
AND-gate assemblies
Despite the sequence errors, all four AND-gate modules were now assembled, so we proceeded to the final assembly steps. For the split-GFP AND-gate the final plasmid design was simple, the C-terminal module would immediately succeed (i.e. exist downstream of) the N-terminal module in the same pSB1C3 plasmid vector. The N-terminal GFP plasmid was restriction digested within the BioBrick suffix, and the C-terminal GFP part (digested in the complementary fashion) was ligated in. This ligation was transformed, verified by analytical restriction digest, and then sequenced. Sequencing using both VF2 and VR primers confirmed the final assembly of this split-GFP gate in the intended order, including the prefix sequence errors previously mentioned.
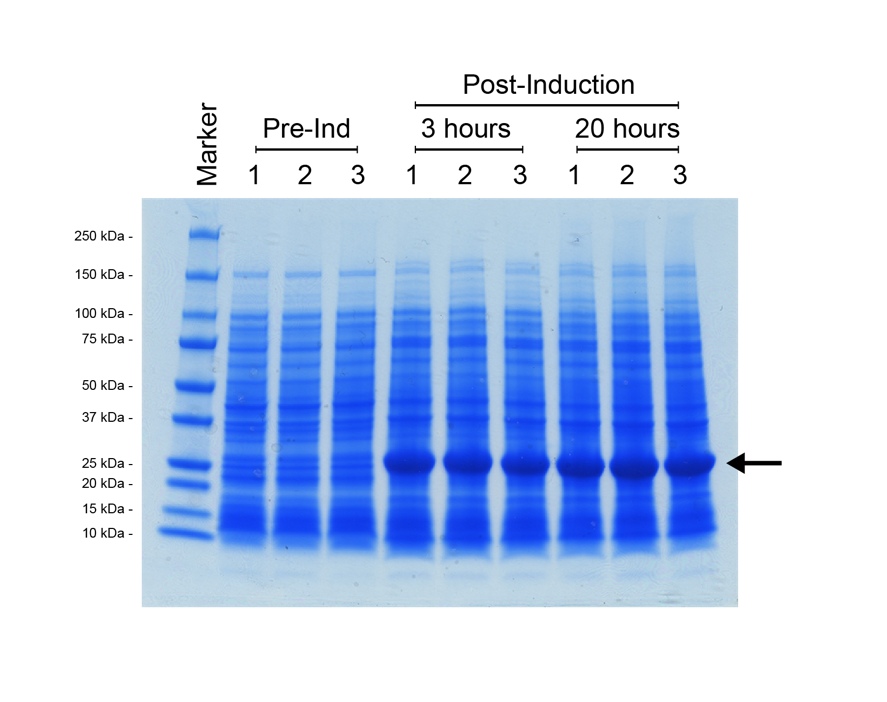
For the split T7 RNAP AND-gate the design of the modular part was the same as for split-GFP, with the T7 RNAP C-terminal module ligated downstream of the N-terminal module, in a pSB1C3 vector backbone. This construct was assembled and verified through digest and sequencing. However, to determine the function of our T7 RNAP gate requires a T7-promoter driven reporter construct. We decided to use the E0040 GFP part as our reporter for split T7 RNAP function. We utilised a preexisting T7 promoter (pT7) construct from the BioBrick registry, [http://parts.igem.org/Part:BBa_K1321338 K1321338]. This part contains the T7 promoter followed by the [http://parts.igem.org/Part:BBa_B0034 B0034] strong ribosome binding site, so that any protein coding sequence could be ligated immediately downstream for expression at high levels when combined with T7 RNAP. We ligated E0040 downstream of [http://parts.igem.org/Part:BBa_K1321338 K1321338] in a pSB1C3 vector, then verified the new composite part by restriction digest and sequencing. We decided to do an additional functional test of this pT7-GFP reporter construct by transforming the plasmid into an E. coli strain that possesses a controllable T7 RNAP integrated into its genome; BL21<DE3>. In this strain, T7 RNAP expression is induced by addition of the lactose analogue IPTG (Isopropyl β-D-1-thiogalactopyranoside). We prepared three overnight cultures of our BL21<DE3> with the pT7-GFP reporter strain, then diluted the overnight culture into 30 ml of fresh L-Broth with chloramphenicol antibiotic, and grew the cultures at 37°C (shaking 225 rpm) until their OD600 = 0.5. At this point a 1 ml sample of each culture was removed, then IPTG was added to each flask for a 1 mM final induction concentration. The flasks were returned to 37°C shaker overnight for 20 hours, with 1 ml samples taken at 3 hours post-induction, and 20 hours post-induction. These cell samples were prepared for SDS-PAGE analysis as discussed in the methods section Figure 6.
It was possible to verify that the expression construct functioned due to the colour of the E. coli cell culture, which became progressively more green over the incubation. However, the SDS-PAGE shows definitively that there has been strong expression of a protein with a molecular weight of roughly 25 kDa, which correlates to the predicted ~27 kDa of the [http://parts.igem.org/Part:BBa_E0040 E0040] GFP. In addition, the gel shows that maximum expression of the reporter was reached within 3 hours, with a negligible visible increase beyond that at the 20-hour time-point.
To finalise our pT7-GFP expression construct, we moved the BioBrick construct from pSB1C3 into another vector – pSB3K3, a kanamycin resistance plasmid with a different origin of replication. This vector move was necessary to allow for both the split T7 RNAP AND-gate and pT7-GFP reporter constructs to be maintained on separate plasmids within a single E. coli cell.
AND-gate testing
Split GFP
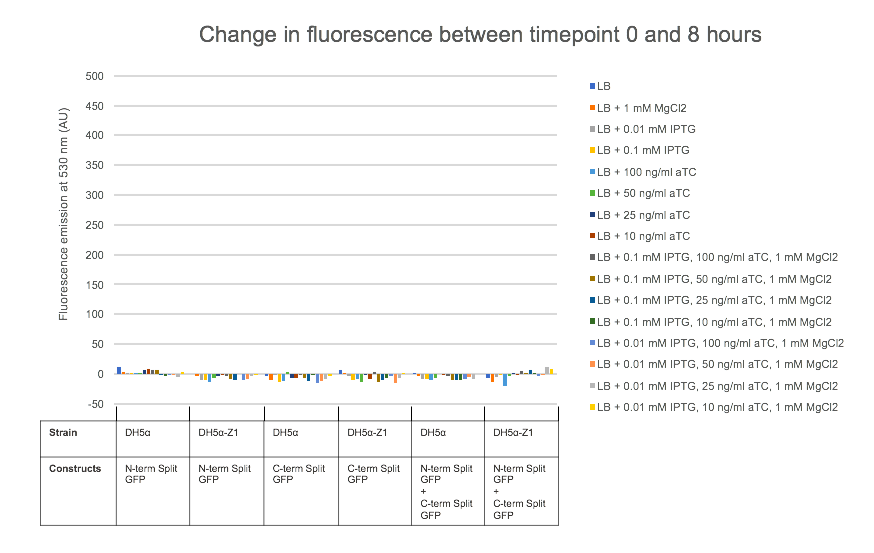
For testing the final split-GFP AND-gate construct K2442408 was moved from E. coli DH5α to the strain DH5α-Z1. As described in the Methods section strains table, DH5α-Z1 contains an overexpressed version of the lacI gene, along with constitutively expressing tetR. These two genes allow for strict control of the lacI and tetR regulated promoters, which should be tightly repressed until the addition of their respective inducer molecules; IPTG and aTC (Anhydrotetracycline). Overnight cultures were prepared of the N-term, C-term, and Full Split-GFP AND-gate constructs; each in both DH5α and DH5α-Z1 strains. The following day these cultures were diluted 1 in 100 with fresh L-Broth, prepared along with a range of inducer and co-factor final concentrations. These conditions are detailed in Figure 7. 200 ul of these culture and inducer combinations were placed into wells of a clear-based black-sided 96 well plate. The prepared plate was then incubated shaking at 300 rpm within a BMG FLUOstar Omega plate reader, where cell growth (absorbance at 600 nm) and fluorescence (excitation at 485 nm and emission at 530 nm) readings taken at 1-hour intervals for 8 hours. 1 mM MgCl2 was added to the dual-inducer inductions as a cofactor which has been shown to improve the reliability of tetR-regulated promoter responses. As shown in Figure 7, no significant change in fluorescence between time 0 and 8 hours was detected in the full AND-gate construct, nor the individual modules. The fluorescence emission reading at 530 nm for the hour 0 timepoint was subtracted from the subsequent reading at the 8-hour timepoint. No noticeable difference in fluorescence can be observed between DH5α or the Z1 variant. A fluorescent response would have been expected in any of the 8 conditions containing both IPTG and aTC, in either strain, but this was not observed.
Cell growth data was also analysed as with Figure 7, which showed that all cultures grew as would be expected, reaching a stationary growth phase by the 8-hour timepoint (Data not shown).
Split T7 RNAP
To test T7 RNAP AND-gate, the same assay protocol for split-GFP gate was followed. In addition to DH5α and DH5α-Z1 carrying N-term module alone, C-term module alone, or the complete AND-gate, the testing strains carried both the pT7-GFP reporter construct in pSB3K3 vector. The same sample preparation and induction conditions were used, along with identical plate-reader settings and time-course. As with the split-GFP AND-gate, the split T7 RNAP AND-gate reported no increase in fluorescence under any condition. While cellular growth measured by absorbance at 600 nm behaved as would be expected.
Discussion
The lack of fluorescent output from either the split-GFP AND-gate or the split T7 RNAP AND-gate can be explained by the errors found during DNA sequencing of the constructs. For the GFP AND-gate, both the N and C-terminal modules possessed improper spacing between the ribosome binding site and their ATG start codon. This error in the C-terminal module was down to a mistake during design of the Forward PCR amplification primer, where the “long” BioBrick prefix was used instead of the two-nucleotide shorter version intended for parts beginning with ATG. With the N-terminal split-GFP module we relied on a pre-existing BioBrick that also contained an elongated prefix; ending TAGA instead of TAG. This may be explained if the original producers of the part amplified it using a Taq PCR polymerase, that adds single Adenines to the 5’ ends of its amplified fragment. With T7-RNAP gate, we were unlucky by selecting a colony for the N-terminal module that contained a 4-nucleotide deletion within the R0040N promoter sequence. If compared to the part-registry annotations of R0040, these 4 nucleotides are located between the -35 and -10 sites of the promoter. The proper spacing of -35 and -10 sites is crucial for E. coli RNA polymerase recruitment and transcription. Had this sequence error been caught before the next assembly stage, then it would have been possible to sequence a larger number of the promoter-ligation colonies, to find a transformant where the R0040N promoter was fully intact. In all three of these cases, small nucleotide sequence errors outside of the coding region of the parts appear to have rendered them inert. With a few more weeks of lab work, it may have been possible to resurrect both final AND-gate constructs by using PCR mutagenesis to correct the sequence errors in each respective module.
Conclusion
During this subproject, we sought to create and characterise two different forms of genetic AND-gate. It was planned that one of these forms of AND-gate would serve as the logic controller of our Campylobacter biosensor, where two systems developed by other subprojects such as mtlR, araC, or AI-2 would act as the transcriptional controllers for AND-gate modules. Due to DNA sequence errors, we were not able to measure a response from either the split-GFP or split T7-RNAP AND gates. However, all the basic parts described have been submitted to the BioBrick registry and should be available for use by future teams.
References
- ↑ Gilbert, M., Mandrell, R.E., Parker, C.T., Li, J., and Vinogradov, E. (2007). Structural Analysis of the Capsular Polysaccharide from Campylobacter jejuni RM1221. ChemBioChem 8, 625–631.
- ↑ Miller, M.B., and Bassler, B.L. (2001). Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 55, 165–199
- ↑ Studier, F.W., and Moffatt, B.A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130.
- ↑ 4.0 4.1 Shis, D.L., and Bennett, M.R. (2013). Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc Natl Acad Sci U S A 110, 5028–5033.
- ↑ Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B.C., Remm, M., and Rozen, S.G. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res 40, e115.