Contents
Overview
Quorum sensing is used by many types of bacteria including Escherichia coli (E. coli) and Campylobacter. The system naturally used by E. coli which involves the LsrR protein binding to and repressing the LsrA promoter (PlsrA) was utilised in this subproject. I constructed a plasmid with E. coli PlsrA followed by green fluorescent protein (GFP). This promoter should be turned off in the presence of the chromosomal encoded LsrR protein. When Autoinducer-2 (AI-2) is present, it binds to the LsrR protein and stops it from repressing PlsrA, allowing PlsrA to start transcription of GFP. I expected the results to show an increase in fluorescence when the plasmid containing PlsrA was introduced into cells which make AI-2, such as DS941 E. coli, when compared to the same construct in cells that don’t make AI-2, such as DH5α E. coli. The results did not show an increase in fluorescence when comparing cells not exposed to AI-2 to cells exposed to AI-2.
Background
We designed our Campylobacter biosensor in the form of an AND-gate, which requires two positive inputs to yield an output signal. In our design one of these input signals would be the presence of the rare sugar xylulose (see mtlR subpage) from the Campylobacter cellular capsule. For the second input signal we intended to exploit the secretion of quorum-sensing molecules, AI-2, by Campylobacter. Our final biosensor device would only report an output signal of GFP fluorescence if both inputs (Xylulose AND AI-2) were present, which would reduce the possibility of false-positive results.
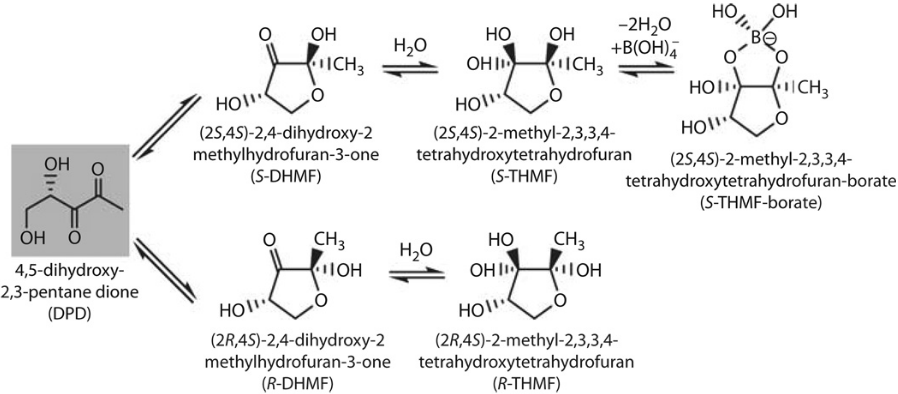
E. coli, and many other bacteria including Campylobacter, use quorum sensing to sense population density and coordinate gene expression dependent on this. Signalling molecules, such as AI-2, are constitutively produced by bacteria. AI-2 is a furanosyl borate diester which can be recognised and produced by many gram-positive and gram-negative bacteria. AI-2 is a byproduct of the activated methyl cycle which is responsible for recycling S-Adenosyl-L-Methionine (SAM). During this cycle, the LuxS enzyme produces the precursor of AI-2, 4,5-Dihydroxy-2,3-Pentanedione (DPD). DPD then spontaneously becomes cyclic forming AI-2 [1]. Cyclic forms of DPD can be seen in figure 1.
AI-2 is then imported into bacteria by the LsrABCD transporter and is subsequently phosphorylated by LsrK kinase. Transcription of the lsrABCD operon is repressed by the LsrR repressor, a DNA binding protein that binds to PlsrA. However, phosphorylated AI-2 is bound by LsrR changing its conformation so that LsrR no longer binds to DNA. Thus phosphorylated AI-2 induces expression of the lsrABCD operon [3].
Previous iGEM teams have used a similar process to detect AI-2 before. The Singapore 2008 iGEM team used a sequence from Rahnella aqualitis which was characterised by researchers in 2000 [4]. This promoter was tested by the Tokyo 2011 iGEM team who discovered it did not work as intended. They then redesigned the promotor using a sequence of the lsrA promoter from E. coli [5]. LsrR is encoded just upstream of the lsrABCD genes in E. coli, and the region between lsrR and lsrA contains the promoter and regulatory sequences for the lsrABCD operon. However, further research found that the Tokyo team missed out the last 14 bases of the promoter sequence which are important for LsrR binding. Therefore, the same sequence that the Tokyo team used was ordered from IDT along with that sequence with the 14 based included on so that the whole LsrR binding site could be tested.
Three different promoters were ordered from IDT. The nucleotide sequence of the whole intergenic region between the lsrR gene and the lsrA gene can be seen in figure 2.
- LsrR promoter - this region can be seen between the { } brackets.
- LsrA promoter - this region can be seen between the [ ] brackets. This is exactly the same sequence that was ordered by the Tokyo 2011 team.
- LsrA+14 promoter - this region can be seen between the second set of { } brackets. This includes the sequence ordered by the Tokyo 2011 team with the last section of the promoter is included.
- lsrR-lsrA intergenic region - this is the full sequence of nucleotides seen in figure 2.
Aims
- Transform PlsrA, PlsrA+14, and lsrR-lsrA intergenic region promoters into plasmids in E. coli.
- Test these plasmids within two different cells; DH5α and DS941. DH5α does not make AI-2 because there is a frameshift mutation in the LuxS gene [6] and DS941 does make AI-2. This allows for comparison of the constructs when AI-2 is present and when it is not.
Materials and Methods
Ultramer Oligo Annealing
The ultramers, PlsrA and PlsrA+14, were 97 and 111 nucleotides long respectively. They were ordered from IDT as single stranded DNA. The annealed ultramers were designed to have sticky ends compatible with EcoRI and SpeI for ligation upstream of other biobrick parts.
- 40µl TE buffer added to the forward and reverse single stranded ultramer to resuspend the ultramers. These were vortexed and left for 10 minutes then vortexed again.
- 10µl of the forward ultramer from step 1 and 10µl of the reverse ultramer from step 1 were added to 80µl of TE buffer.
- This mixture was then left in an 80℃ water bath for 10 minutes for the strands to anneal.
- The annealing reaction tube and roughly 200ml of water was scooped out of the water bath in a beaker and left to cool to room temperature overnight.
PCR Amplification of lsrR-lsrA Intergenic Region
The primers ordered from IDT to amplify the lsrR-lsrA intergenic region can be seen in figure 3.
- Black: random DNA
- Green: prefix
- Blue: reverse complement suffix
- Pink: forward and reverse primers
The template for the PCR was colony PCR of DH5α genomic DNA. The PCR products were analysed by gel electrophoresis, gel extracted then digested at EcoRI and SpeI for ligation. This was then ran on a gel and gel extracted.
Fluorescence Readings
- Overnights were set up from single colonies of strains containing PlsrA promoter construct.
- These were cultured in L-broth with antibiotic at 37℃ shaking at 225rpm.
- The following morning, 200µl of each culture was placed in a well of a 96-well plate.
- GFP fluorescence was measured at excitation wavelength 485 nm and emission wavelength 530nm, along with cell density measured at 600nm, in a FLUOstar omega plate reader.
Results and Discussion
Assembly of PlsrA and PlsrA+14
The PlsrA was tested so the Tokyo 2011 team’s results could be replicated. The PlsrA+14 was examined to see if our predictions of where the LsrR repressor protein binds were correct.
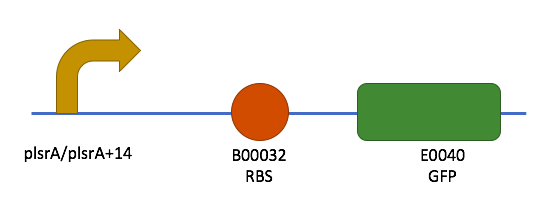
The ultramers containing PlsrA and PlsrA+14 promoter constructs were annealed as described in the methods section. They were then ligated into PSB1C3 in front of B0032 (RBS) and E0040 (GFP). To verify that the promotors had successfully ligated into the plasmids, a unique restriction site, BSU36I, was exploited. This restriction site was unique to the promoter sequences only, which allows insertion into the plasmid to be verified. The results of this restriction digest can be seen in figure 4.
It was expected that the PlsrA and PlsrA+14 in pSB1C3 in front of the RBS (B0032) and GFP (E0040) would be 2941bp and 2955, respectively. The results from the gel image show this to be the length of the plasmid in all lanes. The linearised plasmid runs slightly slower than the uncut plasmid because the uncut plasmid is supercoiled so can run through the gel slightly more quickly than the cut plasmid.
The insertion of the parts was verified in all transformants tested, so they were then verified by sequencing.
- PlsrA followed by B0032 and E0040 were given the biobrick number [http://parts.igem.org/Part:BBa_K2442001 K2442001].
- PlsrA+14 followed by B0032 and E0040 were given the biobrick number [http://parts.igem.org/Part:BBa_K2442003 K2442003].
Assembly of lsrR-lsrA Intergenic Region
The lsrR-lsrA intergenic region was tested to investigate how the whole promoter region, including PlsrR, would act in a plasmid.
The PCR product was made as described in methods. The PCR product was then digested with EcoRi and SpeI so that it could be ligated into PSB1C3. Unfortunately, the PCR product could not be ligated successfully into the PSB1C3 plasmid.
Final Constructs
Overall reporter plasmids made
- PlsrA ligated in front of B0032 (RBS) and E0040 (GFP).
- PlsrA+14 promoter ligated in front of B0032 (RBS) and E0040 (GFP)
The final constructs made can be seen in figure 5.
Construct Testing
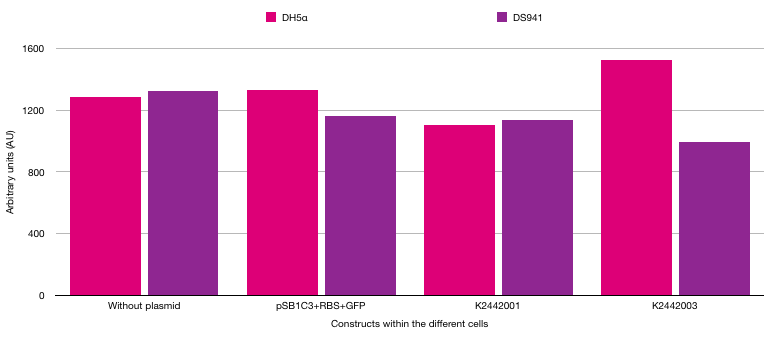
The different strains of E. coli were used to show the difference between fluorescence in cells exposed to AI-2 and cells not exposed to AI-2. DH5α cannot make the LuxS protein as there is a frameshift mutation in the luxS gene [7]. However, DS941 can make the LuxS protein so AI-2 is made in these bacterial cells. If AI-2 is present, the PlsrA should be active and transcribe GFP. If AI-2 is absent, the PlsrA should be off as it will be bound by the LsrR repressor protein. The results from the plate reader can be seen in figure 6.
Discussion
We aimed to make a quorum sensing system to detect the signalling molecule, AI-2. To do this we ordered ultramers for the PlsrA and PlsrA+14 promoters and primers to synthesise the complete lsrR-lsrA intergenic region from E. coli. We then aimed to characterise three promoters; PlsrA, PlsrA+14, and lsrR-lsrA intergenic region using a FLUROstar plate reader and measuring fluorescence emitted from DS941 and DH5α E. coli cells.
There are no results for the lsrR-lsrA intergenic region because we did not succeed in ligating the PCR fragment into the plasmid.
The results were not as expected. It was hypothesised that the cells with PlsrA and PlsrA+14 in DS941 E. coli cells would result in higher fluorescence readings due to GFP being made when the lsrA promoter was activated. However, fluorescence did not increase when comparing a cell with no plasmid to a cell with PlsrA or PlsrA+14 promoters.
Furthermore, there should have been higher fluorescence in DS941 cells compared to DH5α cells. DS941 makes AI-2 which can repress LsrR protein, meaning the LsrA promoter can be activated. However, the results do not show that there was higher fluorescence in these cells. Due to time constraints it was not possible to investigate further. To gain definitive data on the functions of the PlsrA promoter we would have required a larger set of repeated data. In addition, an important future control for this experiment would be using LsrR knockout strains in which PlsrA should be constitutively active.
In future, AI-2 could be bought and added to the overnight culture while E. coli cells are growing to ensure the bacterial cells are exposed to enough AI-2 to bind to the LsrR repressor protein. Another method would be to use one strain of E. coli which produces AI-2 in the culture with another E. coli strain which contains the biosensor plasmid. The E. coli with the biosensor plasmid could detect the level of AI-2 being made by the AI-2 producing E. coli cell.
To fully depress PlsrA, purified AI-2 would need to be added to the cells. However, AI-2 is highly expensive at $400 for 5mg from http://www.ommscientific.com/products.html. For iGEM, a low-cost purification method needs to be developed before an AI-2 based biosensor could be properly tested.
References
- ↑ Vendeville, A., Winzer, K., Heurlier, K., Tang, C. and Hardie, K. (2005). Making 'sense' of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nature Reviews Microbiology, 3(5), pp.383-396.
- ↑ Federle, M. (2011). Autoinducer-2-Based Chemical Communication in Bacteria: Complexities of Interspecies Signaling. Contributions to Microbiology, 16, pp.18-32.
- ↑ Rutherford, S. and Bassler, B. (2012). Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspectives in Medicine, 2(11), pp.a012427-a012427.
- ↑ Seo, J., Song, K., Jang, K., Kim, C., Jung, B. and Rhee, S. (2000). Molecular cloning of a gene encoding the thermoactive levansucrase from Rahnella aquatilis and its growth phase-dependent expression in Escherichia coli. Journal of Biotechnology, 81(1), pp.63-72.
- ↑ Xue, T., Zhao, L., Sun, H., Zhou, X. and Sun, B. (2009). LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Research, 19(11), pp.1258-1268.
- ↑ Wang, L., Forsyth, M., Hullo, M., Merritt, J. and Sperandio, V. (2017). WikiGenes - Collaborative Publishing. [online] WikiGenes - Collaborative Publishing. Available at: https://www.wikigenes.org/e/gene/e/947168.html [Accessed 17 Oct. 2017].
- ↑ Wang, L., Forsyth, M., Hullo, M., Merritt, J. and Sperandio, V. (2017). WikiGenes - Collaborative Publishing. [online] WikiGenes - Collaborative Publishing. Available at: https://www.wikigenes.org/e/gene/e/947168.html [Accessed 17 Oct. 2017].