Contents
Preparation of CaCl2 competent cells
- Dilute 500μl of overnight liquid culture into 20ml of broth with any necessary antibiotics to select for any plasmids already transformed into the cells.
- Incubate at 37⁰C, shaking at 225rpm for 105 minutes.
- Spin down for 2 minutes at 7000G at 4⁰C.
- Discard supernatant, resuspend pellet in 10ml of 50 mM CaCl2, keep on ice.
- Repeat centrifugation for 2 minutes at 7000G at 4⁰C.
- Discard supernatant and the resuspend pellet in 1ml of 50 mM CaCl2, keep on ice.
- CaCl2 competent cells can be kept on ice in the fridge overnight.
Transformation of CaCl2 competent cells
- Add 1μl of plasmid DNA to 100μl of competent cells.
- Heat shock at 42oC for 45 seconds.
- Add 200μl of L-broth of the sample.
- Keep on ice for 2 minutes.
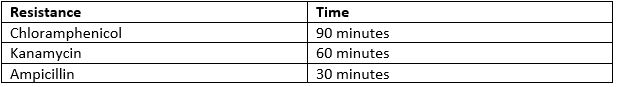
- Incubate the cells at 37oC. The time varies depending on which antibiotic resistance the plasmid holds.
Restriction digests
A 20μl reaction typically contained:
- 2μl buffer
- 4μl Miniprep (or 8μl G-Block) dependant on concentration of Miniprep
- Make up to 20μl with ddH20
- Vortex briefly.
- Incubate at 37oC for at least 60 minutes.
- Heat inactivate restriction digests where appropriate.
Miniprep
- Pipette 1mL of bacterial overnight culture into a microcentrifuge tube
- Centrifuge at 13,000 rpm for 1 min
- Discard the supernatant
- Repeat steps 1-3 two more times
- Add 250μl of P1 buffer and pipette up and down to resuspend the pellet
- Add 250μl of P2 buffer and invert Note: don’t allow this lysis reaction to proceed for more than 5 minutes
- Add 350μl of N3 buffer to neutralise the reaction and invert
- Centrifuge at 13,000 rpm for 10 minutes
- Transfer 800μl of supernatant into a column
- Centrifuge for 1 minute and discard flow-through
- Add 500μl of PB buffer and centrifuge for 1 minute
- Discard flow-through
- Add 750μl of PE buffer and centrifuge for 1 minute
- Discard flow-through
- Centrifuge again for 1 minute to get rid of any residual buffer
- Transfer the column to a microcentrifuge tube
- Add 50μl of EB buffer and let it stand for 1 minute
- Centrifuge for 1 minute
Making a gel
To make a 1.5L buffer:
- Measure 30mL of 50x TAE buffer
- Transfer to a 2L measuring cylinder
- Fill up to 1.5L with distilled water
To make a 1% agarose gel:
- Weigh out 1g of agarose powder
- Transfer to a microwave bottle
- Pour 100mL of your buffer into the bottle
- Microwave until clear
- Cool down to 55⁰C before pouring the gel
Gel electrophoresis
To run the gel:
- Place the gel in the tank and cover it with the buffer
- Load 5μl of DNA ladder into the 2nd well (leave the first one empty)
- Add 5μl of loading dye (30% glycerol, 1% bromophenol blue, 0.5% sodium dodecyl sulphate, diluted in TE buffer) into each restriction digest and pipette up and down a few times
- Load 25μl of each restriction digest into a separate well
- Run gel at 1.5 Amp, 100V for approximately 1 hour
To stain the gel: Ethidium bromide: stain in (concentration) for 40 minutes, destain for 40 minutes, image under UV light
Azure A (for gel extraction): stain in 0.04%/20% ethanol for 15 minutes, destain for 15 minutes (multiple rounds of destaining may be required), image under visible light
Gel extraction using Qiagen kit
- Weigh an empty eppendorf and record the weight
- Cut out a the band from the gel and place it in the eppendorf
- Weigh the eppendorf again and calculate the weight difference
- Add 3 volumes of Buffer QG to 1 volume of gel
- Incubate at 50⁰C for 10 minutes until the agarose has dissolved. Vortex every 2-3 minutes
- Add 1 volume of isopropanol and mix
- Transfer the sample from the eppendorf to a QIAprep spin column in a 2mL collection tube
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough
- Add 500μl of Buffer QG to column
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough
- Add 750μl of Buffer PE and let it stand for 2-5 minutes
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough. Repeat this step
- Transfer the column to a new eppendorf and discard the collection tube
- Add 30μl of Buffer EB to the sample and let it stand for 1 minute
- Centrifuge for 1 minute at 13,000rpm and keep discard the column
Materials and Methods
Condition set up
Sample preparation
- 1
- 2
- 3
Results and Discussion
Outlook
References
- ↑ Kiliç, A. O., Pavlova, S. I., Ma, W. G. & Tao, L. 1996. Analysis of Lactobacillus phages and bacteriocins in American dairy products and characterization of a phage isolated from yogurt. Appl Environ Microbiol, 62, 2111-6.