Described here are the various techniques we used throughout our project so that any other team may reproduce our experiments.
Contents
- 1 Preparation of CaCl2 competent cells
- 2 Transformation of CaCl2 competent cells
- 3 Restriction digests
- 4 Miniprep
- 5 Making a gel
- 6 Gel electrophoresis
- 7 Gel extraction using Qiagen kit
- 8 Ligation
- 9 Annealing oligos
- 10 Polyacrylamide Hydrogel
- 11 Sporulation of Bacillus subtilis
- 12 Difco Sporulation Medium
- 13 Colony PCR
- 14 SDS-PAGE
Preparation of CaCl2 competent cells
- Dilute 500μl of overnight liquid culture into 20ml of broth with any necessary antibiotics to select for any plasmids already transformed into the cells.
- Incubate at 37⁰C, shaking at 225rpm for 105 minutes.
- Spin down for 2 minutes at 7000G at 4⁰C.
- Discard supernatant, resuspend pellet in 10ml of 50 mM CaCl2, keep on ice.
- Repeat centrifugation for 2 minutes at 7000G at 4⁰C.
- Discard supernatant and the resuspend pellet in 1ml of 50 mM CaCl2, keep on ice.
- CaCl2 competent cells can be kept on ice in the fridge overnight.
Transformation of CaCl2 competent cells
- Add 1μl of plasmid DNA to 100μl of competent cells.
- Heat shock at 42°C for 45 seconds.
- Add 200μl of L-broth of the sample.
- Keep on ice for 2 minutes.
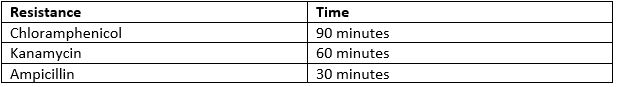
- Incubate the cells at 37°C. The time varies depending on which antibiotic resistance the plasmid holds.
Restriction digests
A 20μl reaction typically contained:
- 2μl buffer
- 4μl Miniprep (or 8μl G-Block) dependent on concentration of Miniprep
- Make up to 20μl with ddH20
- Vortex briefly.
- Incubate at 37°C for at least 60 minutes.
- Heat inactivate restriction digests where appropriate.
Miniprep
We used the protocol from a standard Qiagen QIAprep Spin Miniprep Kit.
- Pipette 1mL of bacterial overnight culture into a microcentrifuge tube
- Centrifuge at 13,000 rpm for 1 min
- Discard the supernatant
- Repeat steps 1-3 two more times
- Add 250μl of P1 buffer and pipette up and down to resuspend the pellet
- Add 250μl of P2 buffer and invert Note: don’t allow this lysis reaction to proceed for more than 5 minutes
- Add 350μl of N3 buffer to neutralise the reaction and invert
- Centrifuge at 13,000 rpm for 10 minutes
- Transfer 800μl of supernatant into a column
- Centrifuge for 1 minute and discard flow-through
- Add 500μl of PB buffer and centrifuge for 1 minute
- Discard flow-through
- Add 750μl of PE buffer and centrifuge for 1 minute
- Discard flow-through
- Centrifuge again for 1 minute to get rid of any residual buffer
- Transfer the column to a microcentrifuge tube
- Add 50μl of EB buffer and let it stand for 1 minute
- Centrifuge for 1 minute
Making a gel
To make a 1.5L buffer:
- Measure 30mL of 50x TAE buffer
- Transfer to a 2L measuring cylinder
- Fill up to 1.5L with distilled water
To make a 1% agarose gel:
- Weigh out 1g of agarose powder
- Transfer to a microwave bottle
- Pour 100mL of buffer into the bottle
- Microwave until clear
- Cool down to 55⁰C before pouring the gel
Gel electrophoresis
To run the gel:
- Place the gel in the tank and cover it with the buffer
- Load 5μl of DNA ladder into the 2nd well (leave the first one empty)
- Add 5μl of loading dye (30% glycerol, 1% bromophenol blue, 0.5% sodium dodecyl sulphate, diluted in TE buffer) into each restriction digest and pipette up and down a few times
- Load 25μl of each restriction digest into a separate well
- Run gel at 1.5 Amp, 100V for approximately 1 hour
To stain the gel:
- SYBR Safe: stain in (concentration) for 40 minutes, destain for 40 minutes, image under UV light
- Azure A (for gel extraction): stain in 0.04%/20% ethanol for 15 minutes, destain for 15 minutes (multiple rounds of destaining may be required), image under visible light
Gel extraction using Qiagen kit
- Weigh an empty eppendorf and record the weight
- Cut out a the band from the gel and place it in the eppendorf
- Weigh the eppendorf again and calculate the weight difference
- Add 3 volumes of Buffer QG to 1 volume of gel
- Incubate at 50⁰C for 10 minutes until the agarose has dissolved. Vortex every 2-3 minutes
- Add 1 volume of isopropanol and mix
- Transfer the sample from the eppendorf to a QIAprep spin column in a 2mL collection tube
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough
- Add 500μl of Buffer QG to column
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough
- Add 750μl of Buffer PE and let it stand for 2-5 minutes
- Centrifuge for 1 minute at 13,000rpm and discard flowthrough. Repeat this step
- Transfer the column to a new eppendorf and discard the collection tube
- Add 30μl of Buffer EB to the sample and let it stand for 1 minute
- Centrifuge for 1 minute at 13,000rpm and keep discard the column
Ligation
For a 10μl ligation reaction (these volumes change be changed depending on the concentration of the gel extracted DNA):
- 3μl of vector
- 5μl of insert
- 1μl of 10x ligation buffer
- 0.5μl of ddH2O
- 0.5μl of ligase
Vortex
For ligating oligo and vector:
- 1μl of oligo
- 5μl of vector
- 1μl of 10x ligation buffer
- 3μl of ddH2O
- 0.5μl of ligase
Vortex
Ligation reactions were left for a minimum of 1 hour or ideally overnight at room temperature or in a temperature-stable room at 16⁰C to ensure maximum ligation efficiency.
Annealing oligos
- Dissolve dried oligo in TE buffer to make a 100μM solution and leave for 10 minutes
To make a 100μl reaction:
- 10μl of top strand 100μM oligo
- 10μl of bottom strand 100μM oligo
- 80μl of TE buffer
- Heat at 86⁰C in a water bath for 5 minutes using a float
- Scoop out some water and the float from the water bath using a large beaker
- Let the water cool down slowly to approximately 30⁰C
- Dilute annealed oligos 1/100
Polyacrylamide Hydrogel
For 10ml of a 5% acrylamide hydrogel:
- 8.3ml PBS Buffer
- 1.7ml 30% Acrylamide/Bis-Acrylamide Solution (37.5:1 weight ratio)
- 100ul APS
- 10ul TEMED
- Add PBS, acrylamide solution and APS to a 15ml falcon tube and swirl to mix thoroughly.
- Add TEMED and again swirl to mix and wait for polymerisation.
For different percentage acrylamide hydrogels, only the volumes of acrylamide solution and PBS buffer need altered.
Sporulation of Bacillus subtilis
Followed the protocol developed by the collaboration between the Bonn and Freiburg iGEM teams from 2016.
- Step 1: Overnight culture- Inoculate a colony of Bacillus subtilis in 4ml LB-medium and let the cells grow overnight at 37°C, 200 rpm
- Step 2: Exponential growth- Measure the OD600 of the overnight culture and dilute by using LB-medium to an OD600 in the region of 0.1-0.2/ml in 10ml LB. Let the cells grow to an OD600 of 0.8/ml at 37°C, 200 rpm
- Step 3: Sporulation- Centrifuge 10ml of the cells at 13,000 x g for 1 minute. Wash the pellet five times with 1x PBS buffer by suspending the pellet in 1ml of 1x PBS and centrifuging. Re-suspend the pellet in 5ml DSM (Difco Sporulation Medium). Let the cells grow for 24 hours at 37°C, 200 rpm.
- Step 4: Lysozyme Treatment- Treat the sample with 416ul of 30mg/ml stock lysozyme, incubate for one hour at room temperature and wash six times with 1x PBS
Difco Sporulation Medium
Fresh DSM was prepared as the FeSO4 rusts when in solution.
For 200ml:
- 1.6g Nutrient Broth
- 0.2g KCl
- 0.2ml MgSO4 (1M)
- 0.2ml MnCl2 (10mM)
- 200ml ddH2O
Autoclave and add:
- 0.5ml CaCl2 (1M)
- 1ml FeSO4 (1M)
Colony PCR
The target gene was first isolated from DH5α cells by colony PCR, where 3 colonies were resuspended in 200µl TE buffer and used as template DNA for 3 separate 50µl PCR reactions. This reduced the chances of only selecting a mutant colony. Conditions were adapted from the NEB PCR Protocol for Phusion High-Fidelity (HF) DNA Polymerase with Standard Taq Buffer[1]for a 50µl reaction, as were guidelines for primer design, annealing temperatures and extension times:
- 5X Phusion HF Buffer (10µl)
- 2mM dNTPs (5µl)
- 10µM Forward Primer (2.5µl)
- 10µM Reverse Primer (2.5µl)
- Template DNA (1µl)
- Taq DNA Polymerase (0.5µl)
- Nuclease-Free Water (31µl)
- Primers were designed to anneal flanking regions of the gene, and to include the biobrick prefix and suffix sequences on either end of the amplified gene. The Melting temperature (Tm) of the primers were to be within 1oC of one another (~60oC), and to have a guanine/cytosine content (GC%) of between 30% and 50%. A 6-nucleotide random DNA sequence was also added to the primers which lets the enzymes cut efficiently.
- A longer initial denaturation time of 5 minutes was used since the DNA was still contained within the cells.
- The annealing temperature was taken to be a 2oC lower than the average melting temperature.
- The extension time was suggested to be 15 seconds per kilobases of DNA being amplified. Following amplification, the PCR products were digested (using a restriction site known to be present in the amplicon) and gel electrophoresis was used to check for successful amplification.
SDS-PAGE
Sodium Dodecyl Sulfate – Polyacrylamide Gel Electrophoresis (SDS-PAGE) was used to analyse the effect of the expression plasmid (designed to overexpress xylose isomerase) on the entire protein complement of the E. coli BL21(DE3) cells, and compare to normal cell protein expression to conclude whether the expression plasmid (part [http://parts.igem.org/Part:BBa_K2442371 K2442371]) works as hypothesised.
- BL21(DE3) strain E. coli cells were first transformed with the expression plasmid. 3 separate fresh 3ml liquid cultures (labelled cultures A, B and C) were made using separate colonies, L-broth, and with the addition of of chloramphenicol (Cm) (1,000X) and glucose (20mM/l). Glucose was added to the media because it precedes lactose in the E. coli sugar-consumption hierarchy, and the lactose-induced lac operon loci is known to transcribe RNA polymerase. As such, a source of glucose suppresses RNA polymerase transcription, and so gene expression only occurs after addition of the lac-operon inducer Isopropyl β-D-1-thiogalactopyranoside (IPTG).
- Each culture was then diluted 100-fold (300µl added to 30ml fresh L-broth) in broth containing Cm, and then was incubated (37oC in a shaking rack at 225 rpm) for a period of 3 hours. During this period, cell growth was measured every hour by optical density (OD), using a spectrophotometer (600nm wavelength). Once each culture reached a pre-induction (Pre) OD of roughly 0.5, an aliquot was removed and kept for analysis during SDS-PAGE as samples showing pre-induction cell protein expression. IPTG was added to induce protein expression in the remaining culture. The cells were then incubated (37oC in a shaking rack at 225 rpm) for another 3 hours, recording the post induction (Post) OD of A, B and C.
- Following protein expression, 1ml of cultures A, B and C were transferred to a 1.5ml eppendorf tube and spun down (13,000rpm for 5 minutes). The supernatant was then pipetted off, and the cells were resuspended in laemmli loading buffer (950µl 5X laemmli sample buffer + 50µl 0.5M dithriotheitol (DTT)). The proportion of buffer added to each culture was normalised to the culture with the highest OD, where that culture received 1ml of laemmli buffer (Table 1).
- The gel used was a 10-well Mini-PROTEAN TGX Precast Gel. 25µl of each of the 6 samples (PreABC and PostABC) were loaded on the gel, along with 20µl of protein marker (Biorad Precision All-Blue Molecular Weight Marker ladder). 25µl of dye was also added to the remaining wells to avoid protein running at an angle. The gel was run until the bands reached the green reference line on the SDS-PAGE kit. The gel was then removed from the precast cassette, and stained with Coomassie blue.