Contents
Overview
Our team aims to develop a functional biosensor to detect Campylobacter jejuni on contaminated surfaces. We aim to engineer E.coli to produce GFP in response to the presence of xylulose – a sugar found in the outer capsule of C. jejuni. This page focuses on utilization of the L-arabinose sensing AraC regulatory protein and its regulated PBAD promoter to construct one of the parts of the final detection system. The regulatory protein AraC binds to operator sequences in the PBAD promoter and activates transcription of downstream genes only when bound to its substrate sugar (L-arabinose). Here we performed saturation mutagenesis on the araC gene to construct AraC mutant library with four randomized ligand-binding pocket amino acid residues corresponding to positions 8, 24, 80 and 82. The AraC mutant library was subjected to fluorescence-based screening using a GFP reporter. Due to unavailability of xylulose we tested for mutants responsive to xylose, decanal or no effector. We isolated and sequenced 4 mutants which showed altered effector specificity. Our results demonstrate an approach that allows to engineer a regulatory protein to respond to an effector molecule of interest and represent steps towards developing a functional biosensor to detect C. jejuni on contaminated surfaces.
Background
Our team aims to construct two alternative regulatory systems as potential components of the biosensor. One of them exploits the MtlR operon, naturally found in Pseudomonas fluorescens. MtlR protein activates transcription of downstream genes upon binding to xylulose. However, this approach may have some disadvantages. Coming from a different organism, the MtlR protein may not function in E. coli cells as expected. Furthermore, the MtlR operon has not been previously exploited in biosensors and may not show sufficient specificity to xylulose. Hence, we aim to construct an alternative regulatory system which exploits the L-arabinose operon.
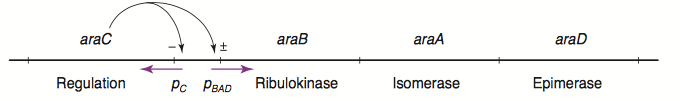
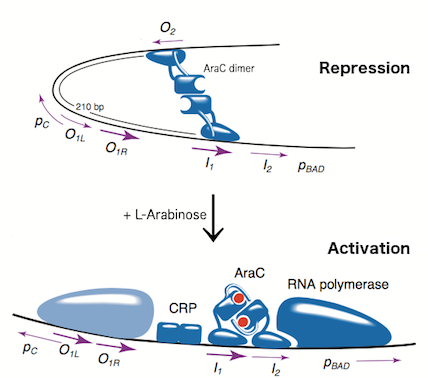
The L-arabinose operon is naturally found in Escherichia coli. The regulatory protein, AraC, acts as a dimer. As depicted in Fig. 1, the operon contains two regulatory cis elements: the PC promoter for the synthesis of AraC, and the PBAD promoter for synthesis of enzymes required for catabolism of L-arabinose. The mechanism of the operon regulation has been described by Schleif et al (2000) and is summarized in Fig. 2. The PBAD promoter contains 3 half sites that each bind to one subunit of AraC - O2, I1, I2. The O1 site is composed of O1L and O1R half sites, which bind both subunits. The O2 half site is within the araC coding region. In absence of L-arabinose the AraC dimer binds to operator half-sites O2 and I1. This causes DNA looping upstream of the PBAD promoter which represses transcription by excluding RNA polymerase from binding to PBAD or PC. Binding of L-arabinose causes a conformational change in the protein such that the DNA-binding domains of the dimer bind to adjacent I1 and I2 half-sites, giving access for RNA polymerase and cyclic AMP receptor protein (CRP) to bind PBAD. This results in transcriptional activation.
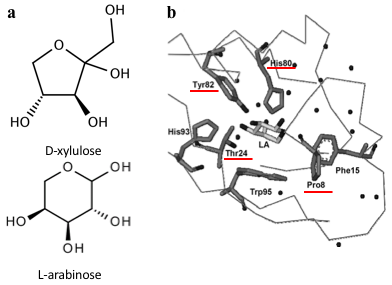
Several previous studies have shown that AraC protein can be engineered to activate transcription in response to non-native small molecules. Some of them include D-arabinose (enantiomer of L-arabinose)[3], mevalonate[4] and triacetic acid lactone[5]. Site-saturation mutagenesis of residues positioned within the ligand-binding pocket of AraC, coupled with fluorescence-based cell sorting, allowed the groups to isolate AraC variants with altered effector specificity. Based on these findings we aimed to use multiple site-saturation mutagenesis and fluorescence-based screening to generate mutant AraC responsive specifically to xylulose. Although xylulose comes in the form of two enantiomers, we chose to focus on D-xylulose, which is the isomer present in the capsule of C. jejuni (Gilbert et al, 2007). Both L-arabinose and D-xylulose are monosaccharides with five carbons, with molecular formulas C5H10O5. As seen in Fig. 3a, arabinose is an aldopentose with an aldehyde functional group at carbon position 1, while xylulose is a ketopentose, possessing a ketone group instead. The 3-dimensional structure of D-xylulose differs from that of L-arabinose, with the most pronounced difference being the 5-membered vs 6-membered heterocyclic ring in its structure. Fig. 3b represents key residues of AraC protein that play a role in ligand binding. Previous studies have shown that mutagenesis of amino acids in positions 8, 24, 80 and 82 is sufficient to change effector specificity[6].
Due to prohibitively high cost of purified xylulose, we aimed to synthesise our own. In the meantime, due to unavailability of the sugar, we screened for mutants responsive to a similar molecule - xylose.
Aims
- Aim 1
To split the AraC regulatory system into two plasmids: regulatory plasmid expressing AraC and AraC mutants and reporter plasmid expressing GFP under control of PBAD promoter.
- Aim 2
To perform multiple site-saturation mutagenesis on the AraC protein at residue positions 8, 24, 80 and 82.
- Aim 3
To isolate AraC variants with altered effector specificity using fluorescence-based screening.
Materials and Methods
Plasmid design
Plasmid construction was as follows.
Oligos containing parts R0011 (LacI-regulated promoter) upstream of B0032 (ribosome-binding site) were synthesised by Integrated DNA Technologies (IDT). Each strand had different, incomplete prefix and suffix sequences, such that when annealed the resulting dsDNA had BioBrick-compatible sticky ends for ligation into EcoRI and PstI digested BioBrick vectors. The resulting product was ligated into high copy number pSB1C3 vector digested with EcoRI and PstI. The ligation reaction was introduced into chemically competent E.coli strain DH5α, and sequence from a chosen colony was verified.
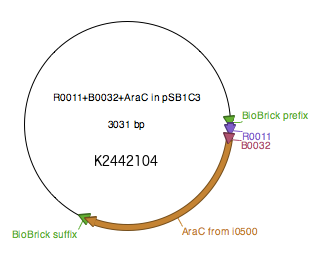
WT araC was amplified from BBa_I0500 using primers araC_BBPre_F and araC_BBSuf_R (see Table 1) to introduce BioBrick prefix and suffix to both ends of WT araC. The PCR product was subsequently digested with XbaI and PstI. pSB1C3-R0011-B0032 was digested with SpeI and PstI. The two digested parts were ligated to generate K2442104 plasmid with WT araC. Sequence was verified.
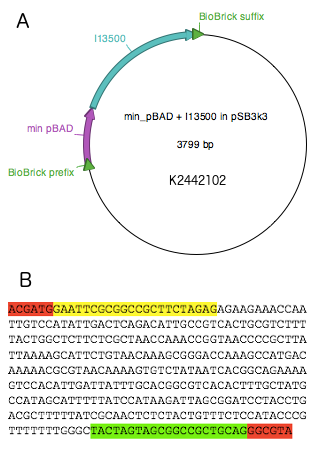
PBADmin was synthesised by Integrated DNA Technologies (IDT) as a G-block and digested with EcoRI and PstI. The cut product was then ligated into pSB3k3 vector digested with EcoRI and PstI. The ligation product was introduced into E.coli strain DH5α by chemical transformation and the sequence was verified. Part I3500 was then digested with enzymes XbaI and PstI and ligated into pSB3k3-PBADmin, digested with SpeI and PstI. This resulted in the final reporter plasmid K2442102, and the sequence was verified.
Library Construction
Primer sequences are listed in Table 1. araC gene was amplified from the BBa_I0500 part using primers araC_F and araC_R. The PCR product was then used in three PCR reactions with the following sets of primers: araC_Frag1_F and araC_Frag1_R; araC_Frag2_F and araC_Frag2_R; araC_Frag3_F and araC_Frag3_R. The PCR conditions were: 98°C for 30s, then 35 cycles of 98°C for 10s, 67.5°C (for F1 and F2) or 59.8°C (for F3) for 30s and 72°C for 1min, then final step of 72°C for 10 mins. The reactions resulted in 3 araC fragments (F1, F2 and F3). Primers araC_Frag1_F, araC_Frag1_R and araC_Frag2_R contain the degenerate sequences NNS at specific triplets so that fragments F1 and F2 contain saturation mutagenesis at codon positions 8, 24, 80 and 82. PCR products were gel purified and DNA concentration of each was measured with a NanoDropTM spectrophotometer. Equimolar aliquots (0.15pmol each) of adjacent fragments were combined (F1+F2 and F2+F3) and PCR-assembled without primers under the following conditions: 98°C for 30s, then 15 cycles of 98°C for 10s, 60°C for 1min and 72°C for 40s, then 72C for 10 mins. 15l of each reaction product was combined and PCR-assembled without primers under following conditions: 20 cycles of 98C for 30s and 72C for 40s. Finally primers araC_BBPre_F and araC_BBSuf_R were added and a final round of PCR was ran with the following programme: 98°C for 30s, then 30 cycles of 98°C for 10s, 60°C for 1 min and 72°C for 40 s, then 72°C for 10 min. QIAQuick PCR purification of the product was performed according to the Qiagen protocol.
The purified PCR product was then digested with XbaI and PstI and ligated into pSB1C3+R0011+B0032 digested with SpeI and PstI. Ligation reaction was ethanol-precipitated, and transformed into electrocompetent DH5α by electroporation. This resulted in the araC mutant library.
Colonies carrying the mutant library were washed off transformation plates with 1.5ml double distilled H2O, (ddH2O) then transferred to a 1.5ml tube. The cell pellet was centrifuged at room temperature, the supernatant was then discarded and pellet resuspended in 1.5ml ddH2O. This centrifugation-resuspension step was repeated 3 times to remove agar plate debris. Plasmid DNA was extracted using the Qiagen Plasmid MiniPrep Kit with 2 of each PB buffer and PE buffer wash steps to ensure maximum extract purity. Sequencing primer VF2 was then added and sample sent for sequencing.
Fluorescence-based screening
Plasmid DNA containing the araC mutant libraries were transformed into DS941 carrying the reporter plasmid K2442102 (in pSB3k3 vector). Transformants were plated on LB agar medium containing chloramphenicol plus kanamycin (to select for K2442102 and K2442104) plus one of the tested inducers: arabinose, xylose or decanal. Concentrations were 40mM for arabinose and xylose and 2mM for decanal. As a control test transformants were also plated on LB medium containing chloramphenicol and kanamycin only. Fluorescence images of the conditional transformation plates were obtained using a GE-Healthcare Typhoon FLA-9500 laser scanner. Excitation was recorded at 473nm, emission was recorded using 520-540nm filter. Colonies which exhibited fluorescence were observed as dark colonies on the scan (Fig. 8). 200 of those which appeared dark on xylose or decanal plates were re-streaked onto plates containing xylose, arabinose, decanal or no additive. Each colony of interest was picked and then immediately short-streaked onto each new condition plate using the same toothpick, into the same position using grids. This method of replica plating ensures the plates can later be aligned and short streaks of the same origin directly compared between the plates. After overnight incubation at 37°C, fluorescence scans of each plate were again obtained using the scan fluorescent scanning method.
Liquid culture fluorescence assay
Colony of interest was inoculated into L-broth containing chloramphenicol and kanamycin. Each liquid culture was grown overnight at 37°C, shaking at 225 rpm. The following day the culture was diluted 1:100 into fresh L-broth containing the above antibiotics plus one of the inducer conditions: 40mM xylose, 40mM arabinose, 2mM decanal. 200l of each culture to be tested was placed in a well of a clear-bases 6-well plate, then incubates at 37°C shaking at 300 rpm for 12 hours in a BMG Fluorostar Omega fluorescence plate reader. Fluorescence of each culture was measured at 1 hour time intervals during the culture experiment, using excitation wavelength 485nm and emission wavelength 530nm. Cell growth was simultaneously tracked by measuring optical density at 600 nm.
Results
Regulatory plasmid K2442104 contains wild type or mutant araC gene under control of LacI-regulated promoter R0011 linked to ribosome binding site B0032. This is to enhance control over production of the AraC, so that the growth of bacteria is not affected by overproduction of the protein. Production of AraC can be stimulated by addition of Isopropyl β-D-1-thiogalactopyranoside (IPTG) to the media. IPTG mimics allolactose and prevents LacI from binding to its operator sequences in the DNA, switching on the R0011 promoter. The construct was ligated into pSB1C3 plasmid backbone (Fig. 4).
The reporter plasmid K2442102 contains GFP gene downstream of minimal PBAD promoter (PBADmin) (Fig. 5A). Minimal PBAD contains minimal DNA sequence from the native PBAD that is sufficient to retain its function (Figure 5B). The promoter retains all the operator sites O1, O2, I1 and I2 required for binding of AraC. As these sites lie within the PC promoter and overlap with AraC coding sequence, the minimal PBAD has the araC start codon ATG mutated to AGT to prevent the protein from being made. PBADmin was created and tested with the help of Steven Kane, member of Colloms laboratory. To enable incorporation of PBADmin into a BioBrick-compatible plasmid, we added BioBrick prefix and suffix sequences to either site of the PBADmin sequence (Fig. 5B). Part K2442102 was expected to work as follows. When AraC is activated by its binding substrate, it binds to I1 and I2 half sites within PBADmin and drives expression of GFP, causing the host cell to produce green fluorescence. For the purposes of testing, the construct was ligated into pSB3k3 plasmid backbone. The plasmid has a different origin of replication from that of pSB1C3, therefore both plasmids can co-exist in the same E. coli cell.
The plasmids were all BioBrick compatible and at every stage of cloning were checked with gel electrophoresis and sequencing. K2442102 and K2442104 were then transformed into DS941 cells and plated onto arabinose conditions to check function (Fig. 6). The scan showed that the cells produced GFP even without the addition of IPTG (an inducer for the lacI-regulated promoter). The promoter is therefore leaky – background transcription of araC from K2442104 is sufficient to activate PBADmin on K2442102.
Fig. 6 shows every colony to have high levels of fluorescence on arabinose. This confirms that the araC and PBADmin parts still function without being adjacent to one another.
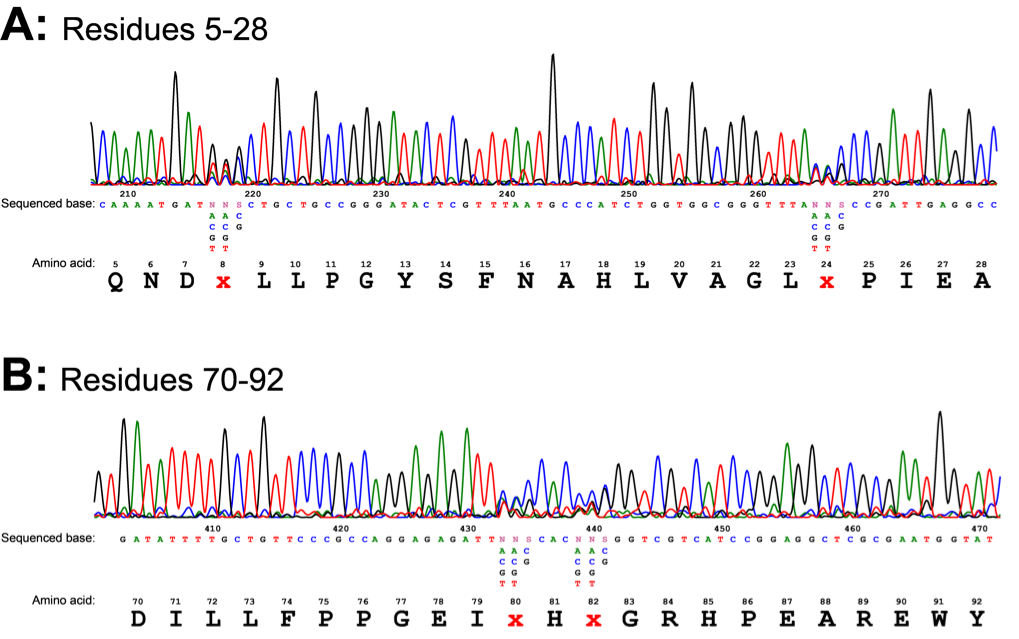
The regulatory plasmid carrying the library of mutant araC was also constructed and transformed as discussed in the methods section. This gave us a few thousand colonies, each containing a binding site pocket variant of the AraC protein. The mutant library DNA was confirmed by sequencing. Sequencing showed the mutations had been introduced at the correct positions (Fig. 7). The trace showed obvious NNS mutations at residues 8, 24, 80 and 82.
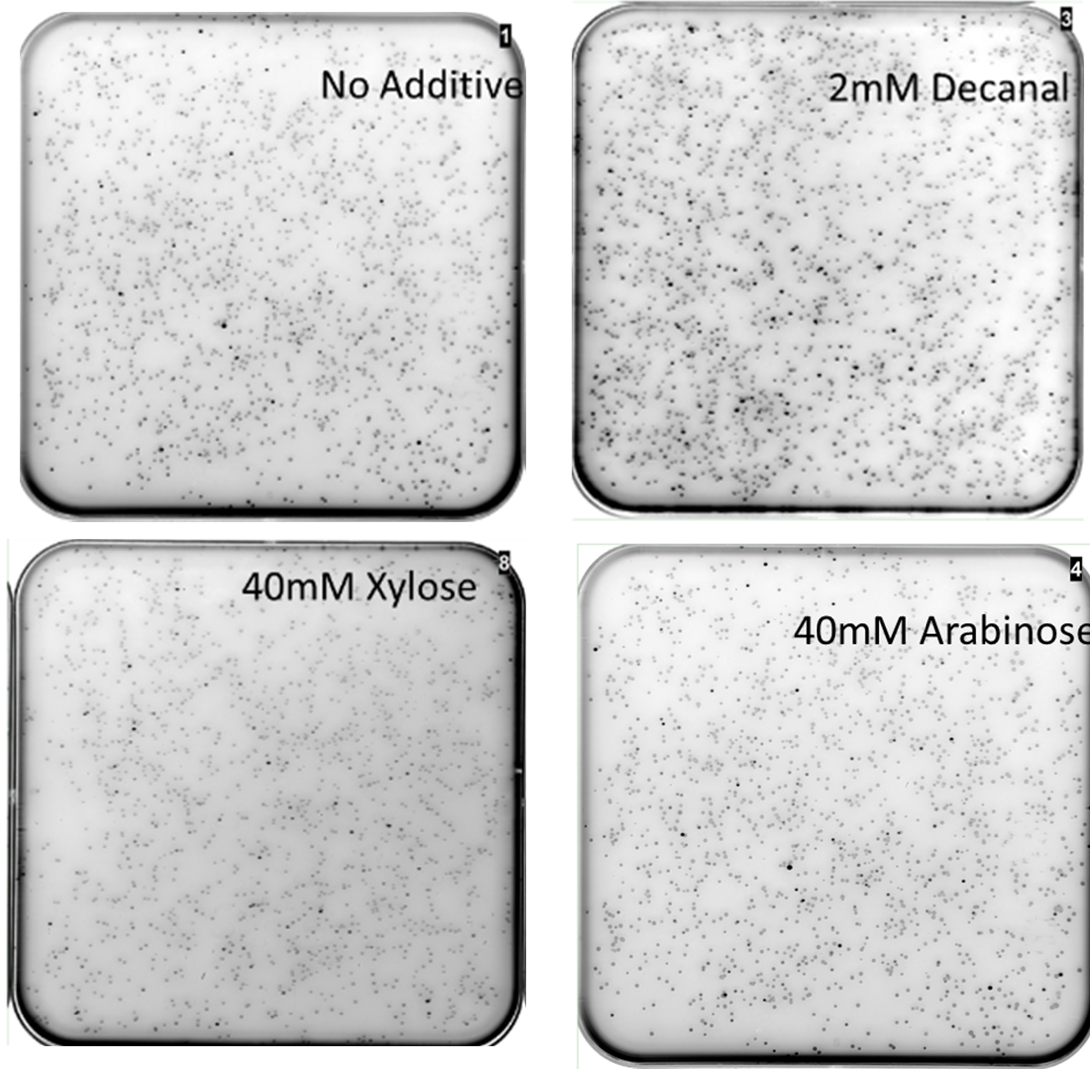
MiniPrep DNA of the mutant araC library plasmid and K2442102 (reporter plasmid) were then transformed into E. coli DS941. These cells were then plated onto LB agar containing kanamycin and chloramphenicol with: A) no additive, B) 40mM xylose, C) 40mM arabinose or D) 2mM decanal and then scanned for GFP fluorescence (Fig. 8). The mutants were screened against no additive to look for mutant AraC with constitutive activity. They were screened against arabinose to check for unmutated/ mutated AraC with wild-type function. Xylose was used as it has a similar chemical structure to xylulose, but was much more economically viable. Xylose does not naturally stimulate WT AraC (Doyle, M., et al., 1972). Decanal is a simple aldehyde, with a 3D structure very different to arabinose. It should have no effect on the binding pocket of wild type AraC, however responsive mutants may be generated. We chose to screen against decanal to check whether mutants obtained are responsive specifically to xylose, or are also inducible at other conditions.
The mutant library was screened in four different conditions. Highly fluorescent, non- fluorescent, and colonies with intermediate fluorescence were seen in all conditions (Fig. 8). Multiple plates of each condition were screened. The presence of non-fluorescent colonies on arabinose shows that AraC has been mutated so that it no longer responds to arabinose. Some fluorescent colonies were observed on plates containing xylose or decanal, suggesting that mutants that responded to these substances might have been obtained. Some fluorescent colonies were even observed with no added sugar (or decanal) in the plate. This shows that the AraC protein has been effectively mutated, as some variations are showing different behaviour to the wild-type protein.
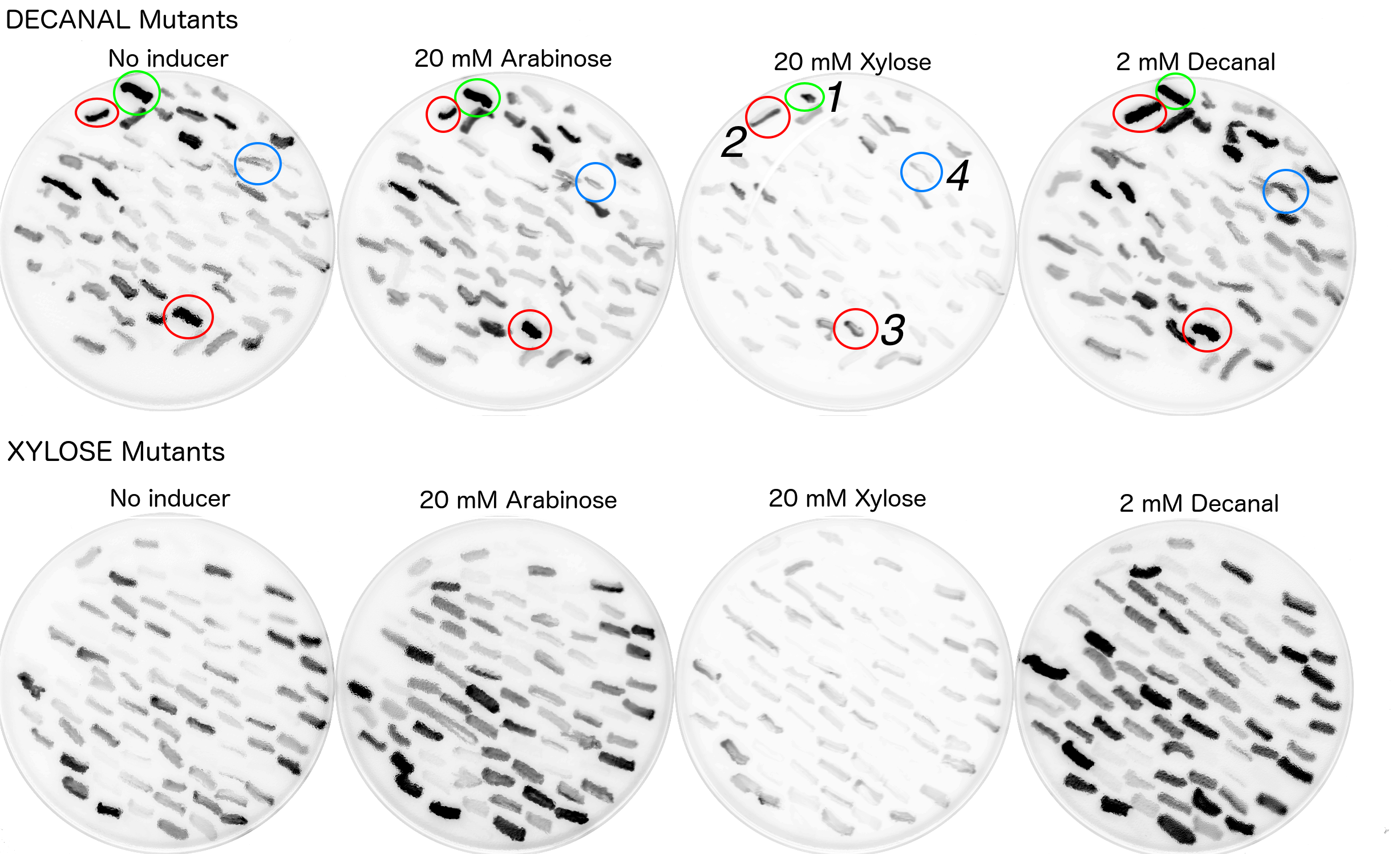
Due to time and labour constraints only around 200 colonies which exhibited fluorescence on either decanal or xylose plates were re-streaked on plates at all four original conditions (i.e. xylose, arabinose, decanal or no chemical). Colonies were picked by referencing a printed out copy of the fluorescence scan and dotting the colonies which appeared to have strong fluorescence. This allowed for room for error. The fluorescence scans of the restreaked mutants showed a variety of behaviours (Fig. 9).
The restreaks (Fig. 9) were examined and four colonies were identified or further examination. All four of these colonies originated from the decanal plate. Colony no.1, (circled in green) was picked as it appeared to be fluorescing under decanal, xylose or arabinose conditions and less so when there was no additive in the agar. Colonies 2 and 3 (circled red) appeared to be fluorescing under all four conditions. Colony 4 appeared to only be fluorescing when streaked onto the plate containing decanal. These four colonies were then picked from the no additive (decanal mutants) plate to be grown in liquid broth (no additive) with antibiotic, overnight. Portions of these overnights were then grown in a plate reader under all four conditions to monitor fluorescence over time (Fig. 10). Biobrick numbers K2442105 - K2442108 were given to the plasmids which contained R0011+ B0032+ a mutant variant of araC. Relevant BioBricks/ composite parts were also monitored for fluorescence to show function when PBAD and araC are split, to reconfirm that our constructs work as intended and to act as controls against the mutant plasmids activity (Fig. 10).
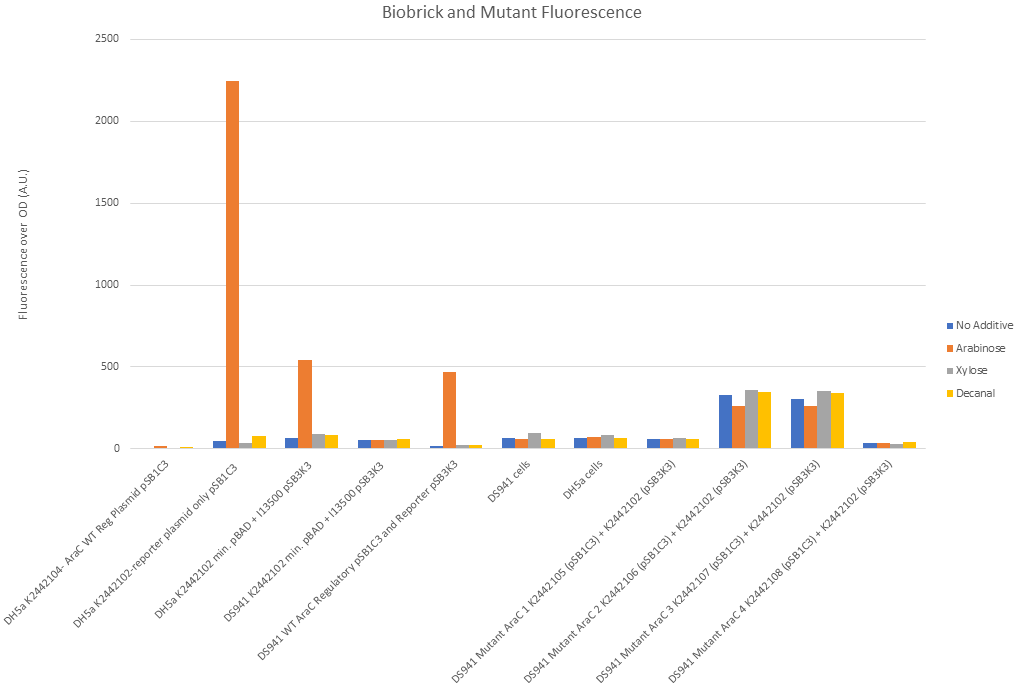
Figure 10 shows that our split I0500 (PBADmin and araC) functions correctly. Reading from left to right on figure 10: There was negligible fluorescence of DH5α cells when transformed with R0011, B0032 and WT araC in pSB1C3. DH5α cells (which have a chromosomal copy of araC) with pSB1C3 with PBADmin and I13500 (second along) showed significant fluorescence under arabinose conditions and negligible fluorescence under all other conditions. The chromosomal copy of araC was sufficient to activate PBADmin and initiated expression of GFP. This reconfirms the functionality of minimal PBAD. Third along in Fig. 10 shows this experiment again, except PBADmin and I13500 are on pSB3K3 instead of pSB1C3. This is a lower copy number plasmid than pSB1C3. Again, expression of GFP was only seen under arabinose conditions, however levels of fluorescence were much lower. This suggests that the change in fluorescence is due to the copy number and must be taken into account in further experiments. The next set of data shows DS941 cells containing PBADmin and I13500 in pSB3K3. DS941 does not have a chromosomal copy of araC. There were negligible levels of fluorescence in this instance, showing that araC is necessary for our construct to express GFP. 5th along shows DS941 cells transformed with both the regulatory plasmid (R0011, B0032+ WT araC in pSB1C3) and the reporter plasmid PBADmin + I13500 in pSB3K3). As they are DS941 cells, the only araC protein present is the one supplied from our transformation. The results were comparable to data set 3, when a chromosomal copy was used to activate PBADmin(on pSB3K3), with high levels of expression only when exposed to arabinose. This shows that the sequence used for wild-type araC in our construct is functional. Empty DS941 and DH5α cells were also scanned for fluorescence - the levels were very low and have been used as a base level for negligible fluorescence.
DS941 cells with the mutant variations of araC in the regulatory plasmid and the reporter plasmid were tested in a separate experiment due to lack of space in the reader. Protocol was carried out in the same way.
Mutant 1, K244105, showed levels of fluorescence comparable to empty DS941 cells. There was no obvious difference between arabinose, decanal and no additive (Fig. 10) when liquid culture was examined. Mutant 2, K2442106 and Mutant 3, K2442106 both showed increased levels of fluorescence compared with empty DS941 cells (Fig. 10). Levels of fluorescence were increased under all conditions, even when there was nothing added to the broth. Arabinose conditions showed the lowest levels of fluorescence for K2442106, suggesting that arabinose may be having an inhibitory effect of this variant of AraC. Mutant 4, K2442108, shows no significant fluorescence (Fig. 10). Any residual fluorescence shown is probably due to E. coli natural fluorescence. It appears to be inactive with no additive and with the addition of the three other molecules.
None of the mutants shows a spike in expression under arabinose conditions. This shows that the AraC molecule has been successfully mutated and is not behaving as the wild-type would do.
The mutant plasmids were also sequenced. AraC naturally has proline at residue 8, threonine at residue 24, histidine at 80 and tyrosine at 82. Colony 1 (K2442105) has glycine at residue 8, no change at residue 24, proline at 80 and tryptophan at 82. Colonies 2 (K2442106) and 3 (K2442107) showed the serine at residue 8, aspartic acid at residue 24, isoleucine at 80 and alanine at 82.
Colony 4 (K2442108) did not show mutations at the specific sites but rather a large portion of the coding region of araC missing.
Discussion
We have successfully shown that the I0500 Biobrick can function when split into two component parts. Figures 5 and 10 show that the wild-type variant of araC behind promoter R0011 and ribosome binding site B0032 on a high copy number plasmid is sufficient to bind to our minimal PBAD promoter and drive transcription of GFP encoded on a low copy number plasmid. The DS941 strain of E. coli does not produce its own copy of the AraC protein, therefore the only AraC which may be binding to the PBADmin promoter is the one on our construct. The component parts have also been shown to work in Fig 10.
Sequencing results confirmed that there the protocol described did cause mutations at residue positions 8, 20, 24, and 80. The four mutants showed different behaviour to the wild-type AraC. The altering of these residues to change the function of AraC was previously done by Tang et al (2008) to generate AraC responsive to the enantiomer D-Arabinose as opposed to L-Arabinose. Naturally AraC does not respond to D-arabinose. L-xylose was chosen to investigate as it has an identical chemical formula to L-arabinose but a different structural formula, just as L-arabinose and D-arabinose do. Decanal is an aldehyde molecule, just as arabinose, however 3D structure of the molecule is very different, allowing for investigation into the response of mutant AraC against different types of molecules.
K2442105 showed a low fluorescence response under all conditions. This shows that the mutations present in this mutant variant of AraC do not show specific activation by any of the conditions tested. However, this was a limited number. The binding pocket has been mutated to the extent where it is no longer responsive to arabinose.
K2442106 and K2442107 showed very high levels of fluorescence under all conditions tested. This shows that it is active not only when arabinose is present. In fact, both showed arabinose conditions to be causing the lowest level of GFP expression, suggesting that arabinose may be actually having an inhibitory effect on mutant AraC. The sequencing results of K2442106 and K2442107 show specific amino acid changes at residues 8, 24, 80 and 82. These have altered the binding pocket it is active without binding anything or possibly that it is activated by a compound present in the cell or abundant in the LB broth/ LB agar. The change in protein structure and activity would need to be greater examined before we could infer how these changes in the binding pocket of AraC are causing this response. The chances of us fining two mutants with the exactly same DNA sequence are very low. This was presumably due to human error, a mislabelling of MiniPreps or the accidental picking of the same colony twice. However, this does reconfirm the activity of this mutant sequence, which was inadvertently replicated and showed the same behaviour.
K2442108 showed negligible expression of GFP. A large portion of the coding region of araC was not present in this mutant, which likely caused the non-functional response.
Any changes between behaviour on the original restreak plates and the fluorescence plate reader were probably due to varying densities of E. coli on the restreak plates. This made the colony appear darker and thus more fluorescent.
All the mutant colonies were only loaded into the plate reader once per colony per condition. These would need to be repeated at very least in triplicate before any more solid conclusions can be made. Also, if time permitted we would have gone on to use error prone PCR on our mutants to optimise their activity, as was done by Tang et al (2008). It must be noted that 200 is an extremely low number of colonies to do initial screening on. Our protocol was expected to have generated 160,000 different amino acid combinations, and the initial transformation generated an excess of 24,000 colonies. Ideally, we would have screened mutants into the thousands to attempt identifying ones which showed a specific response. Unfortunately, this was not possible in the time frame of the project. Screening with a FACS machine, as was done in a previous study[7] , would allow for screening of more mutants. It would also be useful to test different concentrations of each additive on the mutant colonies to investigate if the behaviour is concentration-dependent. As a further experiment mutant cells could be screened against many different molecules such as general sugars, aldehyde molecules or even something not like arabinose at all.
Outlook
The production of the mutant library of AraC is useful for synthetic biology. A catalogue of many of these mutants could possibly make the construction of regulatory systems such as biosensors much easier, and could be applied as a component of a biosensor toolbox. This could allow to generate operons responsive to any molecule of interest, which do not exist in nature.
The protocol undertaken in this investigation did have a reasonable degree of success and could be replicated by other iGEM teams. Time constraints were the main reason for the limited degree of testing. This could be overcome by other iGEM teams by following our protocol of ethanol precipitating the ligation DNA before transformation and recruiting more people to do screening work, or ideally using a FACS machine to sort the many variant mutant colonies.
References
- ↑ Schleif, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends In Genetics 16, 559-565..
- ↑ Schleif, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends In Genetics 16, 559-565..
- ↑ Tang, S., Fazelinia, H., and Cirino, P. (2008). AraC Regulatory Protein Mutants with Altered Effector Specificity. Journal Of The American Chemical Society 130, 5267-5271.
- ↑ Tang, S., and Cirino, P. (2010). Design and Application of a Mevalonate-Responsive Regulatory Protein. Angewandte Chemie 123, 1116-1118.
- ↑ Frei, C., Wang, Z., Qian, S., Deutsch, S., Sutter, M., and Cirino, P. (2016). Analysis of amino acid substitutions in AraC variants that respond to triacetic acid lactone. Protein Science 25, 804-814.
- ↑ Tang, S., Fazelinia, H., and Cirino, P. (2008). AraC Regulatory Protein Mutants with Altered Effector Specificity. Journal Of The American Chemical Society 130, 5267-5271.
- ↑ Tang, S., Fazelinia, H., and Cirino, P. (2008). AraC Regulatory Protein Mutants with Altered Effector Specificity. Journal Of The American Chemical Society 130, 5267-5271.