Contents
Overview
When designing our biosensor device, we wanted to use a dual-input system for the detection of Campylobacter jejuni in order to reduce false positive readings. One of these inputs was a genetic circuit with an inducible promoter - therefore, allowing transcription to be induced by the sugar xylulose. Xylulose is a six carbon sugar which is present on the coat of Campylobacter jejuni, but on few other bacteria. This page describes the process we undertook to build the genetic circuit and also details the tests we carried out in order to characterise parts from the mannitol operon of Pseudomonas fluorescens.
Background
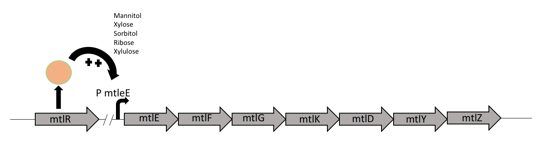
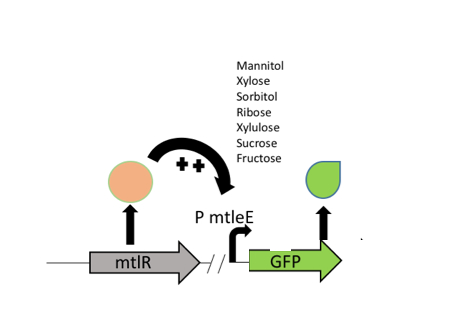
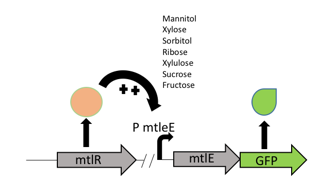
The mtlR subgroup project involved the exploitation of the mannitol operon from P. fluorescens [1] combined with a reporter construct to detect the presence of C. jejuni. Mannitol is a readily available sugar alcohol which is used by many organisms in nature. P. fluorescens, a gram-negative bacterium commonly found in water and soil, can utilize this sugar as its sole energy source as it has organised its mannitol utilization gene set into an operon to transport and convert mannitol (Figure 1). Hoffman and Atlenbuchner [2] described the MtlR regulator from Pseudomonas as a multi-sugar transcriptional regulator (Hoffmann and Altenbuchner, 2017). [2]
Previous studies[2] showed that these sugars switch on transcription of a cluster of genes which are involved in the transportation and metabolism of mannitol. In particular, the mannitol operon "repressor" (MtlR) activates transcription at the mtlE promoter in the presence of polyol sugars, such as mannitol, arabitol or glucitol[2]. In the absence of the sugars, the protein acts in a repressive manner, ensuring no transcription from PmtlE.
The above figure shows the layout of the mannitol operon. MtlR protein is expressed from the upstream mtlR gene and initiates transcription at the mtlE promoter in the presence of certain sugars. Downstream genes involved in sugar transport and conversion are then transcribed.
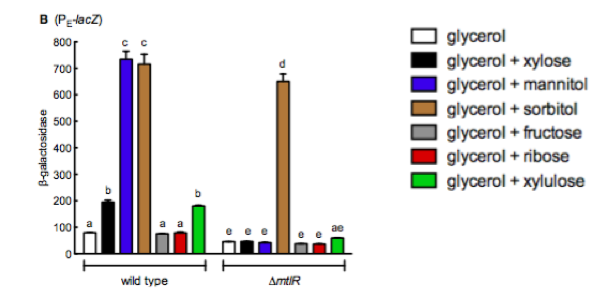
Liu et al (2015) [1] studied the induction of the operon through the sugar xylose, an aldopentose isolated from plants. However, their study found that xylulose (the first breakdown product of xylose) was in fact the sugar that induced the operon rather than xylose. This study expressed lacZ from the PmtlE promoter on the chromosome of P. aeruginosa . Liu et al [1] then measured beta-galactosidase (encoded by lacZ) activity in the presence of different sugars. They used a substrate for beta galactosidase that became fluorescent when cleaved by LacZ, and measured fluorescence in a plate reader. The sugars tested included sorbitol, mannitol, xylose, ribose, fructose and arabinose. In the absence of sugar it was demonstrated that the MtlR protein functions as a repressor and not an activator. Only upon induction via the sugar xylulose, PmtlE became active and was able to induce lacZ expression.
We decided to use this information in construction of our Campylobacter biosensor – it was shown in Gilbert et al (2015) [3] study that xylulose is a component of the bacterial coat of Campylobacter specifically but very few other bacterial strains. Therefore, upon exploitation of the PmtlE promoter and the MtlR transcriptional activator and using xylulose as a trigger, we can infer the presence of Campylobacter.
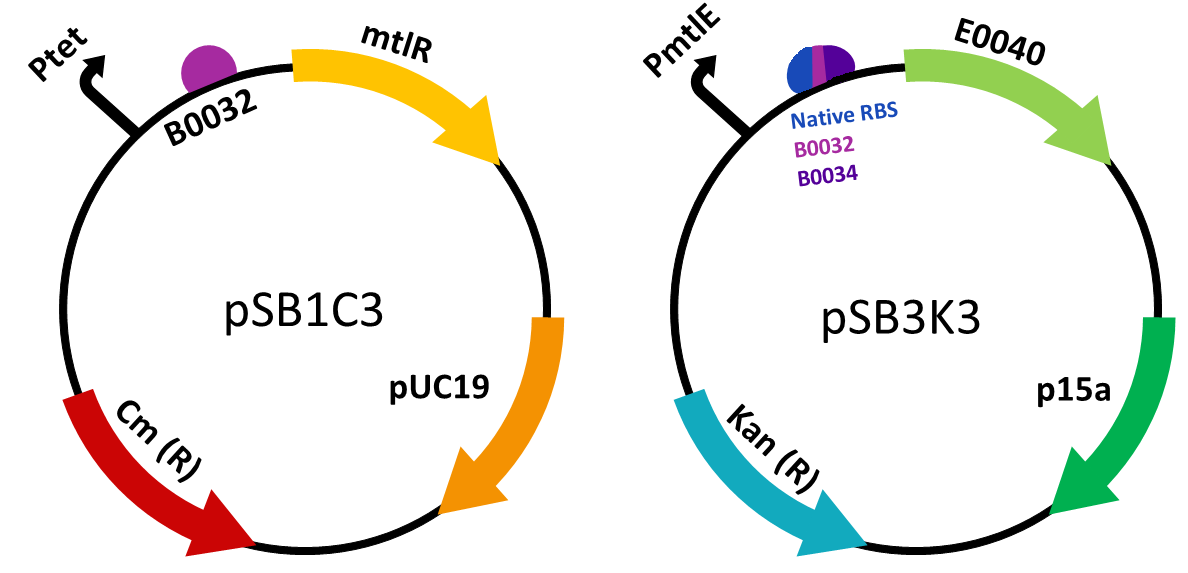
Our biosensor used a two plasmid system to show the interaction between MtlR and xylulose. The first plasmid used the tetracycline regulated Pltet promoter (BBa_R0040) to selectively express the mtlR gene. Upon transcription and translation, the MtlR proteinis then available to interact with xylulose. The second (reporter) plasmid contained the mtlE promoter and a downstream GFP gene. In a finished biosensor device, a higher level of fluorescence of GFP would indicate the presence of xylulose in a sample through the interaction of the MtlR-xylulose complex with the mtlE promoter, driving transcription of the gfp gene.
The mannitol regulatory system will be combined with the Autoinducer-2 quorum sensing regulatory system to create a two part system that will provide a reliable result in the presence of xylulose. With the addition of an AND gate, we created an all or none system, whereby both inputs - xylulose AND autoinducer-2 - must be present to provide a positive result e.g. GFP fluorescence after swabbing a potentially contaminated surface.
Aims
- Aim One
To utilize parts of the P. fluorescens mannitol operon to create a system that will identify the presence of xylulose from campylobacter
- Aim Two
To create a regulatory plasmid expressing mtlR and reporter plasmid expressing mtlE and GFP.
- Aim Three
To study the sugar induction of our constructs and test the viability of these in detecting different sugars
Materials and Methods
In order to test the activity of our mtlR regulatory system under different sugar induction conditions we studied the basal levels of GFP fluorescence using a 96 well plate in the FLUOstar omega plate reader (BMG). Each plasmid was grown in an overnight culture of LB with appropriate antibiotics for each plasmid (chloramphenicol and kanamycin) and then diluted 1:100 with fresh LB and placed into a black bottomed 96 well plate. All readings were done in the plate reader at 37°C shaking at 200 RPM. GFP excitation and emission levels were read, excitation at 485nm and emission at 530nm, every hour for 8 hours.
A standard collection of protocols were followed by all subgroups in the lab and have not been described in detail here.
Results and Discussion
Plasmid Assembly:
We constructed two plasmid constructs- the regulatory plasmid responsible for the transcription and translation of the MtlR protein, and the reporter plasmid which would allow for a measurable response to the MtlR-xylulose interaction. mtlR coding sequence was ordered as a G-block construct from IDT and was ligated into pSB1C3. As this was to be under the control of a tet promoter, the tet promoter and medium RBS (B0032) were ordered as oligos from IDT and were ligated into a separate pSB1C3 plasmid. In order for the mtlR sequence to be behind the promoter and RBS, mtlR was restriction digested from its plasmid and ligated into the promoter plasmid. This produced our final regulatory plasmid.
The reporter plasmid was constructed by first ligating the mtlE promoter into pSB1C3. We ordered the sequence Liu et al (2015) [1] used, which contained the native promoter and RBS. We also decided to experiment with a variety of RBS in order to find the optimal one for our genetic circuit. We used the mtle promoter ligated to two different RBS, one stronger (B0034) and one weaker (B0032). These variants were ordered as single-stranded oligonucleotides, annealed together (to make double-stranded oligonucleotides) and then ligated into pSB1C3. We then used GFP from the kitplate (Biobrick number – E0040). This was restriction digested out of the plasmid and ligated behind Ptet and the various RBS.
In order to test in the presence of both plasmids, the gene construct present in the reporter plasmid was built in pSB1C3 and then digested and ligated into a new plasmid, pSB3K3. This ensured both plasmids had a different origin of replication and therefore could both be present in one cell. It also ensured we could select for cells which contained both the plasmids required to carry out testing.
Testing Constructs:
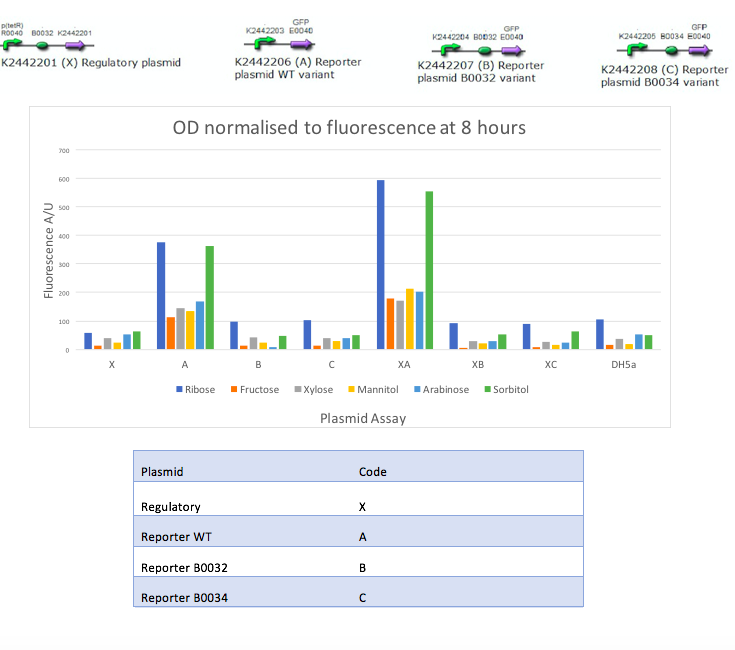
Our aim was to replicate the findings of Liu et al [1] and as such we studied a number of sugars and recorded GFP output in the presence of different inducers. To replicate their results, each reporter variant was tested with each sugar and the reporter variants and regulatory plasmid were tested together with each sugar. These sugars were selected based on their paper [1] and other similar sugars available in the lab were used. We tested these combinations with 6 different sugars; mannitol, sorbitol, sucrose, xylose, ribose, and fructose. Our controls were DH5α empty cells, fresh LB, and each plasmid alone. The two constructs were individually tested with a variety of sugars in order to record the fluorescence in the absence of MtlR. Controls were set up without the constructs in order to measure the basal level of GFP fluorescence. Our results showed:
Discussion
Liu et al (2015) [1] carried out similar experiments to those conducted in this project utilizing B-galactosidase as a reporter to look at the levels of gene expression after inducing the mannitol regulator. They studied the induction of the reporter plasmid in a wild type system and in the absence of mtlR. Their results, figure 4, show around a 7 fold increase in B-galactosidase expression in the presence of mannitol and sorbitol in the wilt type, whilst the other sugars they tested with, xylose, fructose, ribose and xylulose show little increase in response. Upon removal of mtlR, the B-galactosidase response in the presence of mannitol noticeably drops to the levels of the other sugars, however, sorbitol continues to induce the system.
The results obtained from our tests do not reflect the findings of Liu et al [1]. Whilst they demonstrated a strong response to sorbitol and mannitol, our constructs showed ribose and sorbitol evoke the largest fluorescence response across all plasmids. A potential reason our results do not reflect those of Liu et al is the use of different organisms. Our findings could show that E.coli exhibits a different response to ribose and mannitol than Pseudomonas fluorescens. E.coli may not utilize these sugars in induction of this system and therefore downstream genes are not turned on as they are in pseudomonas.
The use of the reporter plasmid alone with varying sugars was to show the requirement of mtlR in this construct. However, figure 3 shows a relatively strong reporter response from the reporter plasmid alone. Although the GFP gene is present it lies downstream of the mtlE promoter, which is activated by the MtlR protein in the presence of sugar. In this experiment, the sugar is available however there is no MtlR protein to bind to and activate the promoter and therefore GFP should not be expressed (Fig 5). As the regulatory plasmid does produce a fluorescence response in the absence of the regulatory protein, this suggests that E.coli may have its own regulatory system that can induce the reporter of this construct. E.coli may produce its own protein that can regulate the induction of the mtlE promoter and therefore produce a GFP response. We expected that only when both the regulatory and reporter plasmids are present, then fluorescence should be shown (Figure 6). Although we get a response from the reporter plasmid alone, when the regulatory plasmid is introduced there is almost a 2 fold increase in GFP response. This indicated that MtlR does induce this system within E.coli and does impact the expression of downstream genes.
Our testing however lacks a no inducer control. This control would allow for the understanding of our system and its induction. As the reporter plasmid alone shows fluorescence response, it may be that this system is constitutively on and therefore will always show some fluorescence response. As our results show an almost doubling of fluorescence with added sugar this also indicates that the sugar is needed to induce a meaningful response, however without this control it cannot be certain that the system isn’t always on.
As the WT reporter shows fluorescence both in the presence and absence of MtlR it would be expected that the other reporter variants, WT mtlE with additional B0032 RBS and WT mtlE with additional B0034 RBS, would show similar results. However, these two variants show very little fluorescence activity. This suggests that upon designing these plasmids, there has been an issue in reducing the activity of mtlE. The mtlE promoter gene sequence was taken from the liu et al paper [1] and adapted in order to add different ribosome binding sites. The lack of activity of these two variants suggests that removing the region we depicted as the WT RBS was an important promoter element, and its removal has compromised the working of the variants. In order to understand this further, future experiments using more variants of this promoter and recording their fluorescent response would need to be conducted.
Outlook
Overall, the testing of our constructs shows much variability to the results obtained by Liu et al [1]. Mannitol did not induce our system whilst ribose did, suggesting difference in sugar response between Pseudomonas and E.coli may have played a part. Upon testing all variants, our results show that wild type reporter produces the greatest fluorescence response, both alone as well as with the regulatory plasmid. As the other variables of the reporter plasmid do not fluoresce once induced with sugar, this suggests the modifications made to mtlE have impacted the function of the promoter and thus changed the ability of the sugar-MtlR complex to bind and induce. Unexpectedly, our reporter plasmid shows a fluorescence response in the absence of the regulatory plasmid, indicating that E.coli may have its own regulatory proteins that can control this system similarly. Future work would include finding and knocking out any potential regulatory molecules that are wild type to E.coli.
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Liu, Y., Rainey, P., and Zhang, X. (2017). Molecular mechanisms of xylose utilization by Pseudomonas fluorescens: overlapping genetic responses to xylose, xylulose, ribose and mannitol.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Hoffmann, J., and Altenbuchner, J. (2017). Functional Characterization of the Mannitol Promoter of Pseudomonas fluorescens DSM 50106 and Its Application for a Mannitol-Inducible Expression System for Pseudomonas putida KT2440.
- ↑ Gilbert, M., Mandrell, R., Parker, C., Li, J., and Vinogradov, E. (2017). Structural Analysis of the Capsular Polysaccharide from Campylobacter jejuni RM1221.